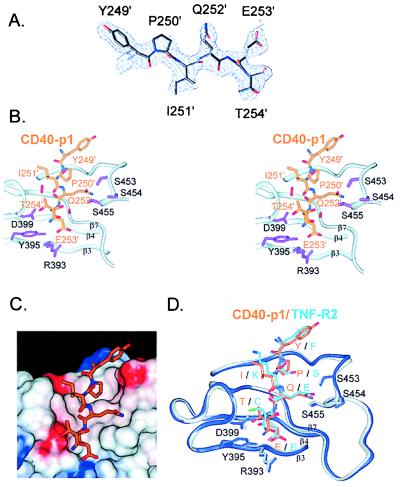
Figure 3.
Receptor recognition by TRAF2. (A) Refined model of CD40-p1 (Tyr-249–Thr-254) superimposed on the solvent-flattened, MAD-phased, 2.4-Å-resolution electron density map (1σ). (B) Stereo view of the TRAF2–CD40-p1 contacts. The TRAF2 backbone is depicted with ribbons. Side chains of TRAF2 residues positioned to make hydrogen bonds to CD40-p1 are shown in purple. The serine tongs, in which conserved serines 453–455 form hydrogen bonds with CD40 Gln-252, are shown at the upper right. CD40 Glu-253 is at the bottom, within hydrogen-bonding distance of TRAF2 Arg-393 and Tyr-395. CD40 Thr-254 is within hydrogen-bonding distance of the conserved Asp-399. Primes (′) denote CD40 residues. (C) CD40-p1 (stick model) shown on the solvent accessible surface of TRAF2 colored by electrostatic potential (−8 to +8 kT/e; blue, positive; white, neutral; and red, negative). The side chains of CD40 Pro-250 and Ile-251 contact a hydrophobic region of the binding cleft, and the remaining CD40 side chains make polar or charged interactions. (D) Comparison of CD-40-p1 (orange) and TNF-R2 (light blue) bound to TRAF2 (CD40-p1 complex, gray; TNF-R2 complex, blue). Single letters denote the residues in CD40–TNF-R2. Three residues in the TNF-R2 peptide not present in CD40-p1 were omitted. The TRAF2 C domains in the two complexes were superimposed without reference to the receptor peptides. TRAF2 adopts similar conformations in the two complexes. The two receptor sequences make distinct side-chain contacts.