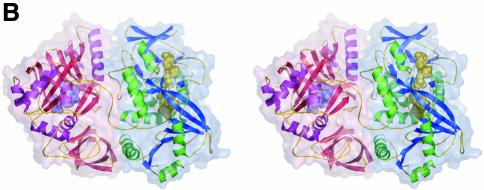
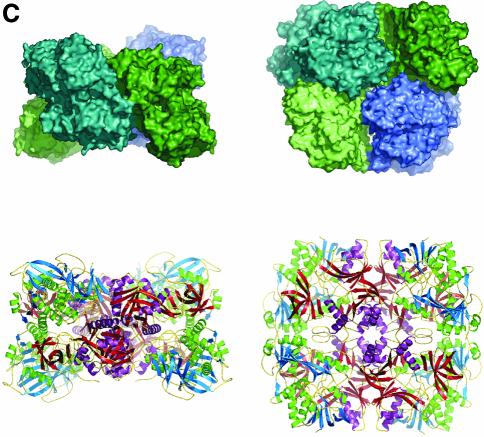
Fig. 1. Structure of DMGO. (A) Alignment of DMGO with human sarcosine dehydrogenase (SHD), human dimethylglycine dehydrogenase (DMGDH) and human T-protein or aminomethyltransferase (AMT). Residues identified as essential to the activity of active site 1 or 2 are shaded in grey. Conserved residues are depicted in bold. The positions of mutations that cause non-ketotic hyperglycinaemia are identified by underlining the corresponding residue in AMT. (B) Stereo view of a DMGO monomer. The FAD-binding domain is represented in blue strands and green α-helices, while the folate-binding domain is depicted in red strands and purple α-helices. FAD and folinic acid are rendered in CPK spheres coloured yellow and blue, respectively. The molecular surface is rendered transparent and coloured according to functional domain: the flavin oxidase domain in blue, and the N5,N10-methylene-THF synthase domain in pink. (C) Structure of the DMGO tetramer. The upper view displays the molecular surface of the tetramer, each monomer represented in a distinct colour; the bottom view displays the backbone in ribbons colour coded as (B). Both orientations are related by a 90° rotation along the horizontal. This and other figures have been generated using the programs Turbo-Frodo (Roussel and Cambillau 1996), Pymol (DeLano, 2002) and GRASP (Nicholls et al., 1991).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.