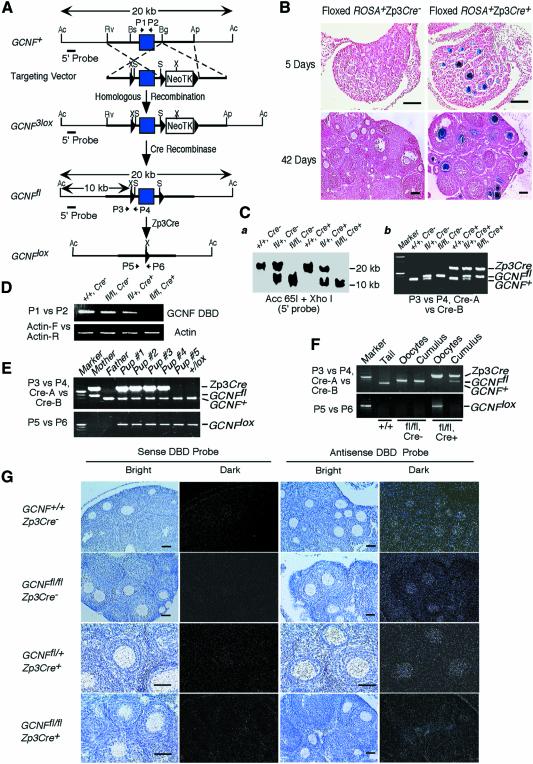
Fig. 1. Generation of an oocyte-specific GCNF knockout mouse model. (A) Structures of the GCNF3lox, GCNFfl and GCNFlox alleles and the targeting vector. LoxP sites, filled triangles; the DBD-encoding exon, filled box. Restriction enzyme sites of Acc65I (Ac), ApaI (Ap), BglI (Bg), BsiEI (Bs), EcoRV (Rv), XhoI (X) and SalI (S) in each allele are shown. Positions of PCR primers (P1 to P6), and the 5′ probe for Southern blot analysis are indicated. (B) Cre recombinase activity on the floxed ROSA transgene in the ovaries determined by β-galactosidase staining. Bar scale, 200 µm. (C) Genotype analysis of tail biopsies. (a) Southern blot analysis showing a 20 kb band for the GCNF+ allele and a 10 kb band for the GCNFfl allele. (b) PCR analysis using primers (P3 and P4) to distinguish the GCNFfl and GCNF+ alleles, and primers (Cre-A and Cre-B) to determine the presence of Zp3Cre transgene. (D) RT–PCR analysis showing the loss of the DBD encoding region of the GCNF mRNA in the GCNFfl/flZp3Cre+ ovary. (E) Genotyping of tail DNA showing the complete deletion of the GCNF DBD encoding exon in the progenies from the cross between GCNFfl/flZp3Cre+ females and wild-type males. (F) PCR analysis showing complete deletion of the GCNF DBD encoding exon in ovulated oocytes, but not in cumulus cells. (G) In situ hybridization showing the loss of the DBD region of the GCNF mRNA in the oocytes of GCNFfl/flZp3Cre+ mice. Bar scale, 50 µm.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.