Figure 2.
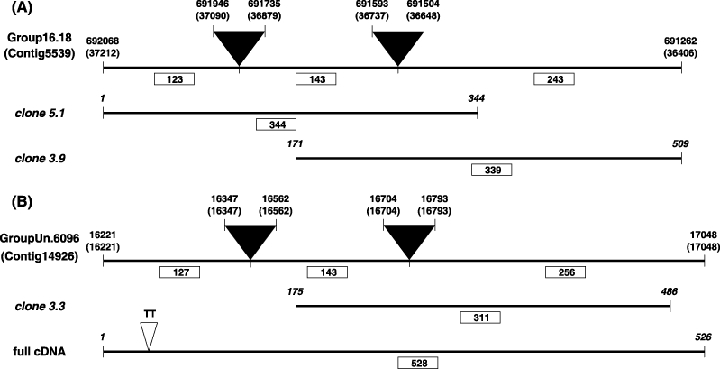
Alignment of two in silico spliced honeybee genome sequences with different cDNA fragments of the bee venom allergen Api m 6. (A) Api m 6 was present in mapped scaffold 16.18 from genome assembly 4.0 (positions marked above the bar, along with coordinates for the corresponding Contig5339). Api m 6 is characterized by two introns (depicted by black triangles; splicing sites given on top) and this haplotype of the genome assembly showed a perfect sequence-level match with cloned 5′- (clone 5.1) and 3′-RACE fragments (clone 3.9). The predicted protein from this sequence is identical to the described Api m 6 variant 1. (B) A second transcript variant was found by cDNA sequencing (full cDNA). This transcript has a nearly identical match to unmapped scaffold GroupUn.6097 (Contig14926), the only difference being a TT-insertion mutation (depicted by a white triangle) in the 5′-UTR. The 3′-end of transcript variant 2 corresponds also to another cloned 3′-RACE fragment (clone 3.3). Fragment lengths (in bp) are given below the bar and in boxes.
