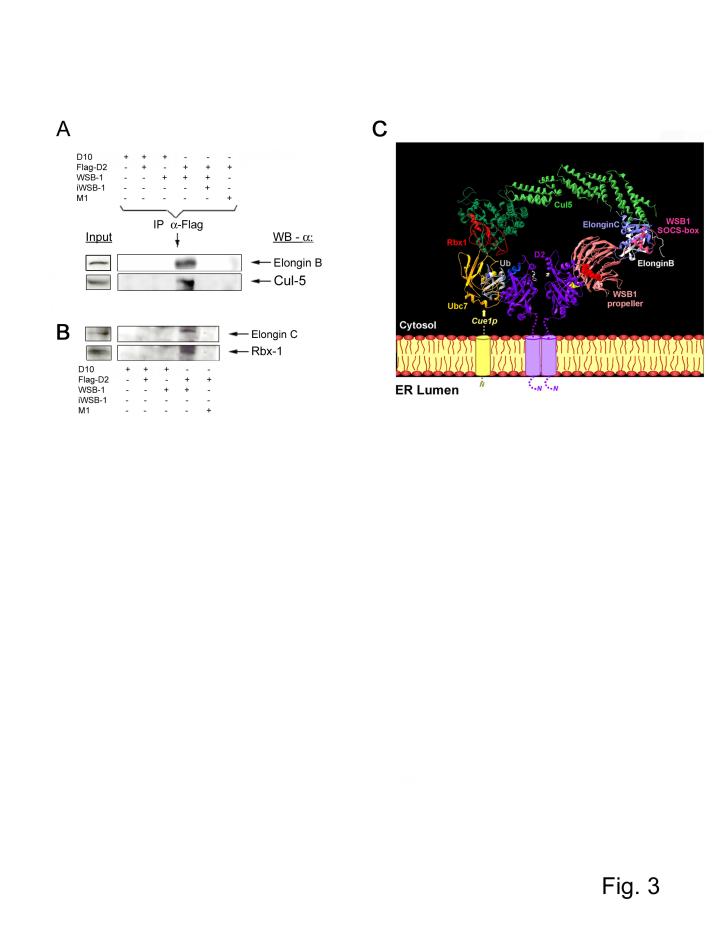
Figure 3.
Composition of the ECSWSB-1 catalytic core complex. (A-B) Lysates of HEK-293 cells transiently expressing the indicated plasmid combination were pulled down with anti-Flag antibody and pellets were processed for western blotting as indicated. (C) In order to assemble the ECSWSB-1 ubiquitinating complex, Elongin C (pdb 1LM8; 14) was docked on the N-terminal part of Skp1 within the structure of the Cul1-Rbx1-Skp1-Fbox(Skp2) complex (pdb 1LDK, 13) and a model of Cul5, which shares 30 % sequence identity with Cul1, was used. The Cul5-Rbx1-Elongin C-Elongin B-VHL complex was reconstructed using the VHLα domain as template for the SOCS-box motif of WSB-1, because such motifs bind Elongin C 12,13. Despite a similar fold for the F-box of Skp2 and the α-domain of VHL, the binding mode with Skp1/ElonginC is different 20. Only the first three helices of the α-domain (three-helix cluster) constitute the common core between F-box, VHL α-domain, and SOCS-box 27.The TrcP1-β-catenin-Skp1 complex (pdb 1P22; 20) was used as template after superimposition of Skp1 on the Skp1/ElonginC structure of the complex described above. The model of the WSB-1 propeller was superimposed on the structure of the TrcP1 propeller and adjusted manually to join the C-terminus of the propeller to the N-terminal end of the SOCS-box through a predicted 5 amino acid loop in WSB1. In contrast to TrcP1, WSB-1 does not have a long linker, so that the propeller is near to the SOCS-box motif. The N-terminal part of WSB-1, which cannot be modeled at this stage, should correspond to a helical extension, located on the Elongin C side of the propeller. Data on the structure of Cue1p are not available and its interaction with Ubc7 is indicated with a dotted line and an arrow. D2 is represented as a dimer 28 with the T4 near the active center (white). The 18-amino acid loop of the right D2 subunit, which can not be modeled at this stage and for which only the start and end positions are indicated in yellow, is oriented towards WSB1, in which the 3 critical amino acids (R174, R315, Y218) are shown as red spheres 25.