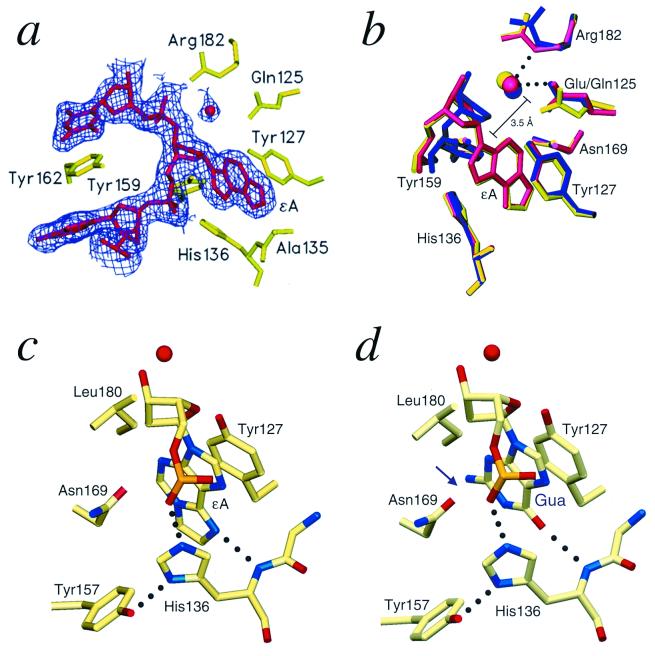
Figure 2.
AAG active-site structure. (a) The 2Fo − Fc electron OMIT density contoured at 2σ above the mean (purple) clearly shows the position of the flipped-out ɛA and a bound water molecule in the active site of the E125Q/ɛA complex. The OMIT density for the wild-type AAG/ɛA-DNA complex has a similar appearance. (b) A superposition of the active sites of the E125Q/ɛA-DNA (green), wild-type/ɛA DNA (red), and wild-type/pyr-DNA (blue) complexes shows these DNAs and the active-site water are bound in similar orientations. However, the pyr ring has rotated to optimize the geometry of a hydrogen bond between pyr N4′ and the bound water. (c) The main chain amide of His-136 makes a key hydrogen-bonding interaction with N6 of ɛA. The N6 of an unmodified adenine would be protonated and repelled by the His-136 amide nitrogen. The side chain of His-136 bridges between the Tyr-157 side chain and the phosphate of the ɛA nucleotide. This fixes the position of the imidazole ring, which stacks against the alkylated base. (d) A guanosine modeled in the active site by superposition on the ɛA nucleotide reveals a clash between N2 of guanine and the Asn-169 side chain (arrow). This steric clash and the conformational constraints on the Asn-169 side chain are best visualized by examining the atomic coordinates of the crystallized complexes (PDB ID codes 1ewn, 1f4r, and 1f6o).