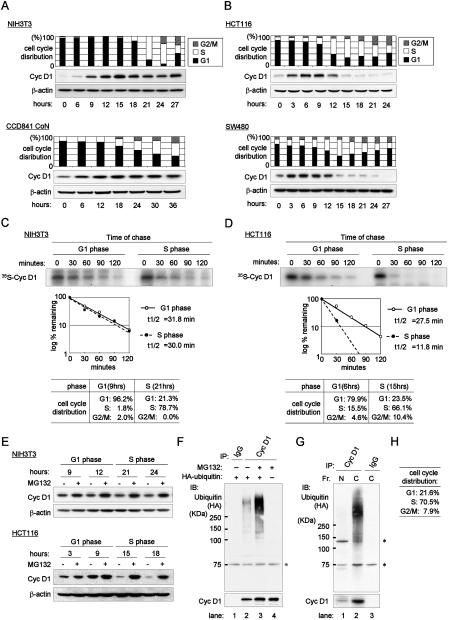
Figure 1.
Cyclin D1 is destabilized during S phase through the ubiquitin-proteasome pathway in cancer cells. (A, B) Expression profile of cyclin D1 during cell cycle progression in cells released from quiescence. Normal cells (A) and cancer cells (B) were tested. (A) NIH 3T3 mouse fibroblasts and CCD841 CoN normal colon epithelium. (B) HCT 116 and SW480 cells. CCD841 CoN cells were synchronized at G0/G1 phase by treatment with ACL-4 media without EGF for 24 hrs, and then stimulated with complete ACL-4 media containing EGF. Other cells were released from quiescence by serum stimulation. Samples were collected at indicated time points. Cell cycle distributions were determined by flow-cytometry and percentages of each phase are indicated. Western-blots were performed with cyclin D1 and β-actin antibodies, respectively. (C, D) Pulse-chase analysis of cyclin D1 in NIH 3T3 cells (C) and HCT 116 cells (D). Cells were released from quiescence and labeled with 35S-methionine for 1 hour when most of the populations were in G1 phase (9 hrs for NIH3T3 and 6 hrs for HCT 116) or in S phase (21 hrs for NIH3T3 and 15 hrs for HCT 116). The cells were chased with cold methionine for the indicated times and then lysed. Cyclin D1 was immunoprecipitated and analyzed with SDS-PAGE. Autoradiography was performed. Levels of metabolically labeled-cyclin D1 were estimated by quantitative scanning using Quantity One software (Bio-Rad) and blotted on the graph to determine the half-life of cyclin D1. Cell cycle distribution is indicated below the figures. (E) Turnover of cyclin D1 is mediated by the ubiquitin-proteasome pathway. NIH3T3 and HCT 116 cells were released from quiescence by serum stimulation and treated in the presence (+) or absence (−) of MG132 for 2 hrs prior to harvesting at each time point. Western blot was performed with cyclin D1 and β-actin antibodies. (F–H) Polyubiquitination of cyclin D1. (F) HCT 116 colon cancer cells were transfected with HA-tagged ubiquitin cDNA (lanes 1–3) or without it (lane 4) and then synchronized to S phase through the sequential manipulation of serum starvation and stimulation. Cells were treated with 25 µM MG132 (lanes 3 and 4) or without it (lanes 1 and 2) for one hour. Lysates were immunoprecipitated with antibodies to cyclin D1 antibody (lanes 2–4) or control IgG (lane 1) and immunoblotted with a HA antibody (upper panel) or a cyclin D1 antibody (lower panel). Asterisks indicate background non-specific bands (F–G). (G) Nuclear (N) and cytoplasmic (C) proteins were fractionated (Fr.) from cell lysates collected in Panel F, lane 3. Nuclear and cytoplasmic extracts were immunoprecipitated with antibodies to cyclin D1 (lanes 1 and 2) or IgG (lane 3) and immunoblotted with a HA antibody (upper panel) or a cyclin D1 antibody (lower panel). (H) Cell cycle distributions.