Figure 1. The role of the mitochondrial genome in energy generation.
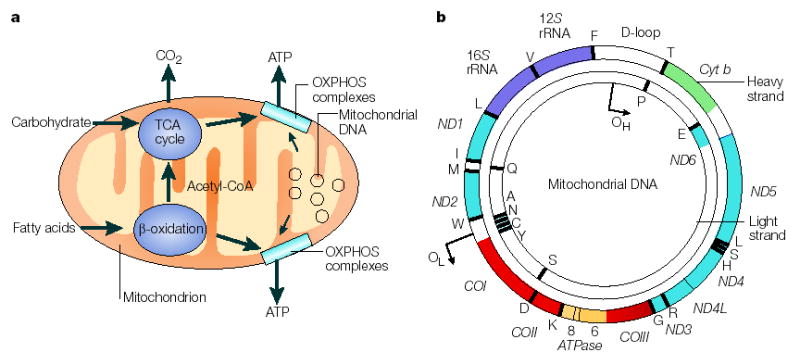
a This highlights the importance of the mitochondrial genome in contributing polypeptide subunits to the five enzyme complexes that comprise the oxidative phosphorylation (OXPHOS) system within the inner mitochondrial membrane — the site of ATP synthesis. The reoxidation of reducing equivalents (NADH (reduced flavin adenine dinucleotide)) that are produced by the oxidation (reduced nicotinamide adenine dinucleotide) and FADH2 of carbohydrates (the tricarboxylic acid (TCA) cycle) and fatty acids (β-oxidation) is coupled to the generation of an electrochemical gradient across the inner mitochondrial membrane, which is harnessed by the ATP synthase to drive the formation of ATP. b A map of the human mitochondrial genome. The genes that encode the subunits of complex I (ND1–ND6 and ND4L) are shown in blue; cytochrome c oxidase (COI–COIII) is shown in red; cytochrome b of complex III is shown in green; and the subunits of the ATP synthase (ATPase 6 and 8) are shown in yellow. The two ribosomal RNAs (rRNAs; 12S and 16S, shown in purple) and 22 tRNAs, indicated by black lines and denoted by their single letter code, which are required for mitochondrial protein synthesis are also shown. The displacement loop (D-loop), or non-coding control region, contains sequences that are vital for the initiation of both mtDNA replication and transcription, including the proposed origin of heavy-strand replication (shown as OH). The origin of light-strand replication is shown as OL.
