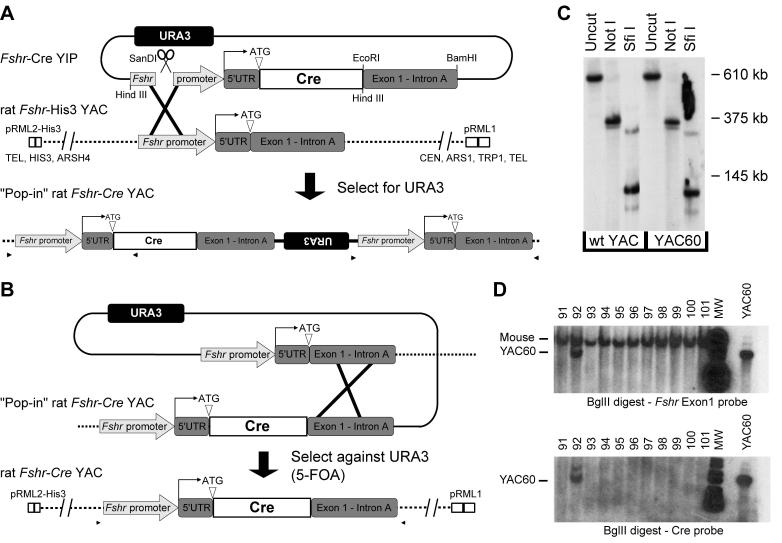
Figure 1.
YAC60 was generated by a two-step replacement via homologous recombination to insert Cre-recombinase at the Fshr translational start site in an Fshr-containing rat YAC. (A) Step one: a yeast integrating plasmid (YIP) with Fshr sequence flanking Cre recombinase was transformed into the YAC-containing yeast and selected for gain of the URA3 marker. Restriction endonuclease sites used to generate the pRS406-Fshr-Cre YIP are noted. (B) Step two: spontaneous revertants with excised YIP vector sequence (i.e.: retaining either the wildtype Fshr or Fshr-Cre sequence in the YAC) were identified by growth in the presence of 5-FOA, which selects against YACs retaining URA3. (C) Limited restriction endonuclease mapping of YAC60. YAC DNA was digested with the noted restriction endonuclease, resolved by PFGE, and analyzed by Southern blot analysis by hybridization with a probe to exon 10. Molecular weight marker migration is shown to the right of the autoradiogram. (D) Southern blot analysis of tail DNA from potential transgenic founder mice was performed using Bgl II restriction digestion and hybridization with probes against exon1 and Cre. Positive transgenic mice were identified by the presence of a faster migrating band (noted in line 92).