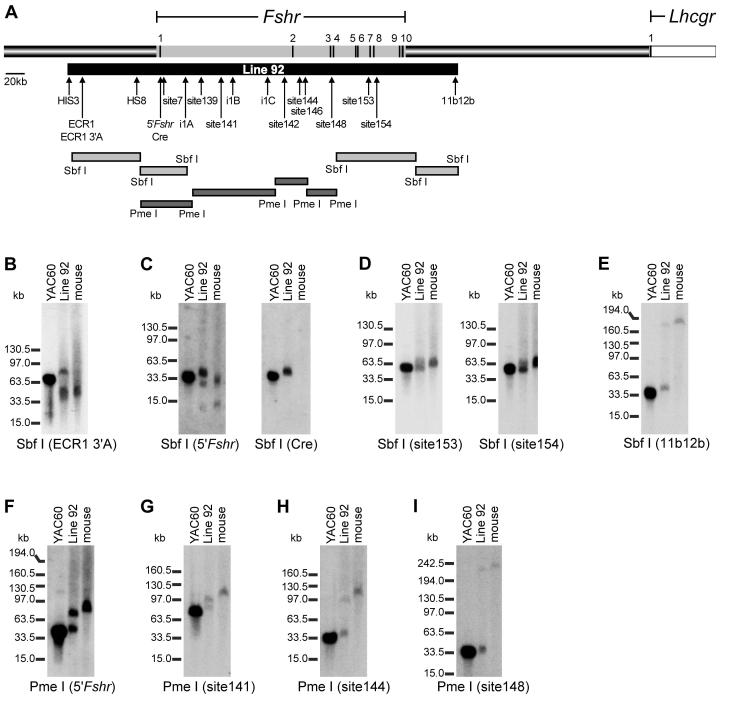
Figure 2.
Mapping of YAC60 in line 92. (A) Schematic representation of 750kb of rat genomic sequence that spans the Fshr coding sequence (grey shading on top bar), including the neighboring gene, LH receptor (Lhcgr, white). Arrows denote the positions of markers (HIS3, ECR1, HS8, Cre, site7, site139, i1A, i1B, i1C, and 11b12b) used for PCR-based mapping of YAC60 in line 92 as well as probes used for restriction endonuclease mapping in Southern blot hybridization analysis (ECR1 3’A, 5’Fshr, Cre, site141, site142, site144, site146, site148, site153, site154, 11b12b). The position of each Fshr exon is noted as a dark line on the grey bar and its number indicated above the bar (note: exon sizes are not to scale). The region of the locus identified within Line 92 is noted by the black bar below, beginning 98kb 5’ to the first transcriptional start site and extending 80kb 3’ of exon 10. Shaded bars indicate restriction fragments used for mapping. Mapping of the YAC60 transgene in line 92 was performed by digestion of high molecular weight DNA with Sbf I or Pme I followed by Southern blot analysis using the indicated probes. In each experiment, digestion of YAC60, line 92, and wildtype mouse DNA were compared. Sbf I digested DNA was analyzed with the following probes: (B) ECR1 3’A probe, (C) 5’Fshr and Cre, (D) site153 and site154, and (E) 11b12b. Pme I digested DNA was analyzed with the probes (F) 5’Fshr, (G) site141, (H) site144, and (I) site148. Restriction endonucleases and hybridization probes are noted below each autoradiogram. Migration of molecular weight standards is noted to the left of each autoradiogram.