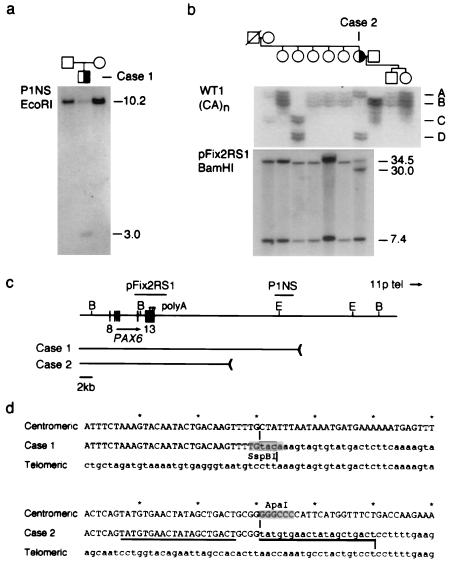
Figure 1.
Identification of 3′ PAX6 rearrangements in sporadic aniridia. (a) Southern analysis of case 1. Probe P1NS detects a novel 3.0-kb EcoRI fragment in the affected child, but not in his parents, and a 10.2-kb EcoRI fragment in normal DNA. (b) Haplotype and Southern analysis of case 2. (Upper) Segregation of the WT1 (CA)n repeat polymorphism. Four alleles are distinguished. Each allele is represented by an ensemble of fragments that differ from the mean length by 1–2 repeat units; these arise by replication slippage during PCR amplification. Case 2 is heterozygous (AD) at WT1. (Lower) Probe pFix2RS1 detects 34.5-kb and 7.4-kb BamHI fragments in normal DNA, and an additional 30-kb BamHI fragment was in the affected female, who is heterozygous. The mutation arose on a paternal chromosome 11 and is linked to WT1 allele D. (c) Restriction map showing PAX6 exons 8–13, polyadenylation sites (arrowheads), hybridization probes, and the centromeric deletion breakpoints in these two cases. The breakpoints are located 22.1 kb and 11.6 kb from the 3′ end of PAX6. E, EcoRI; B, BamHI. Some of the EcoRI sites have been omitted. (d) DNA sequence spanning the centromeric (uppercase) and telomeric (lowercase) breakpoints of cases 1 and 2 in normal DNA and the deletion junctions. The junction points are indicated by vertical lines and the 20-bp duplication in case 2 is underlined. The rearrangement in case 1 creates an SspBI restriction site at the junction.