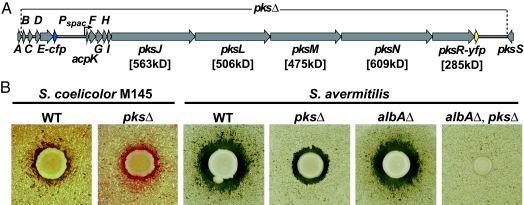
Fig. 1.
Genetic organization and activity of the B. subtilis bacillaene synthase. (A) A simplified schematic of the bacillaene (pks) gene cluster indicating the points of fusion to the fluorescent protein genes. The extent of pks gene deletion in the pksΔ strain and the location of pksE-cfp (blue arrow) and pksR-yfp (yellow arrow) fusions are indicated. In only the cells containing a fusion of cfp to the pksE ORF, genes downstream of pksE-cfp are expressed under isopropyl β-d-thiogalactoside control of the Pspac promoter. Predicted molecular weights of large multimodular proteins are listed below the gene names (genolist.pasteur.fr/SubtiList). (B) Mutations in the individual pks genes or a deletion of the entire pks gene cluster reveal growth and developmental phenotypes in coculture with Streptomycetes (SI Text). (Left) B. subtilis WT and pksΔ cocultured with S. coelicolor M145 on YEME pH7 agar. A red ring (undecylprodigiosin produced by S. coelicolor) is visible around the pksΔ but not the WT strain. (Right) NRPS/PKS activity produced by B. subtilis inhibits S. avermitilis growth in coculture. The activity is lost in a pksΔ strain. The loss of PK activity in a pksΔ strain is obscured by the effect of subtilosin (product of the sbo-alb gene cluster). Total loss of activity is seen by comparison of pks+, albAΔ and pksΔ, albAΔ double mutant strains.