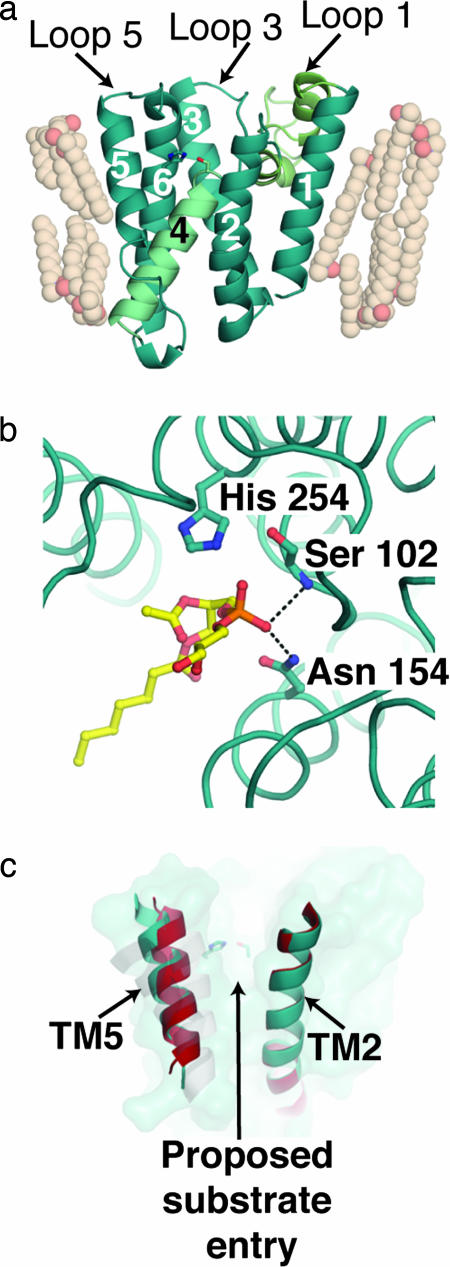
Fig. 1.
Views of GlpG. (a) Molecular architecture of E. coli GlpG [Protein Data Bank (PDB) ID code 2IRV]. TMs are numbered sequentially. TM4, which contains the catalytic residue Ser-201, is colored light cyan; and loop 1, which comprises two unusual helices H1 and H2, is colored light green. (b) Phospholipid binding in the proposed active site of GlpG. Dashed lines indicate hydrogen-bonding interactions to the phosphate group that likely parallel the oxyanion hole stabilization during catalysis. (c) GlpG surface (teal) and superimposed TM2 and TM5 helices from two molecules seen in structure solved by Ben-Shem et al. (red and teal). A grayscale overlay of TM5 for the other structures of GlpG (PDB ID codes 2NRF and 2IC8) is shown.