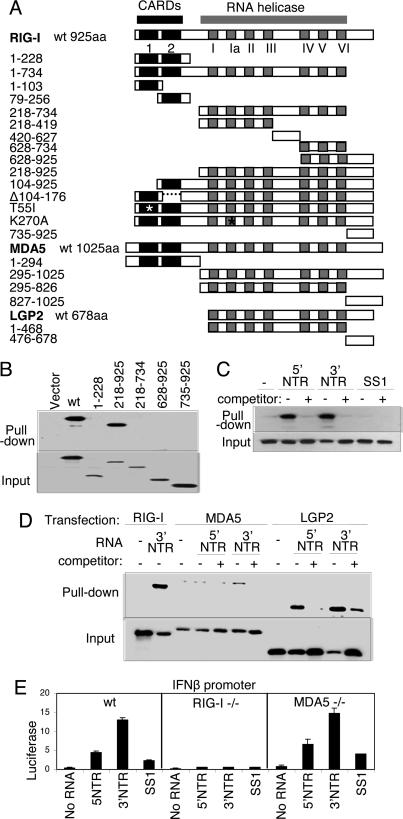
Fig. 1.
Constructs and RNA binding properties. (A) Domain structure of the RIG-I, MDA5, and LGP2 constructs showing the positions of the tandem CARDs and the RNA helicase domain and its subdomains conserved among the helicase superfamily (15). Point mutations are indicated by an asterisk. The amino acid region encoded by each construct is shown at left. (B) Extract from Huh 7 cells that were transfected with empty vector or plasmid expressing Flag-tagged RIG-I constructs encoding WT (wt) RIG-I or the indicated amino acid regions of RIG-I were mixed with pIC-agarose beads and subjected to pull-down assay for dsRNA binding. RIG-I proteins within the input material (Lower) and recovered as pull-down product (Upper) were evaluated by immunoblotting using anti-Flag antibody. (C and D) Cytoplasmic fraction (20 μg) from 293 cells transfected with plasmid encoding Flag-tagged WT RIG-I (C) or with Flag RIG-I wt, Flag-MDA5 wt, or Flag-LGP2 wt (D) were mixed with 1 μg of in vitro-transcribed biotin-UTP HCV 5′ NTR RNA, 3′ NTR RNA, or SS1 RNA alone (−) or with an excess of unlabeled homologous competitor RNA (+). RNA–protein complexes were recovered by pull-down assay using streptavidin affinity gel. Flag-tagged protein within the pull-down fraction or 25% of input material was analyzed by immunoblotting using anti-Flag antibody. (E) wt, RIG-I-null, or MDA5-null MEFs were cotransfected with plasmids encoding constitutive Renilla luciferase and firefly luciferase controlled by the murine IFN-β promoter (2). After 24 h, the cells were mock-transfected or transfected with 10 ng of in vitro-transcribed, gel-purified RNA encoding the HCV 5′ NTR, 3′ NTR, or SS1 region (6). After 24 h, the cells were harvested, and extracts were subjected to dual luciferase assay. Bars show relative luciferase values and SD.