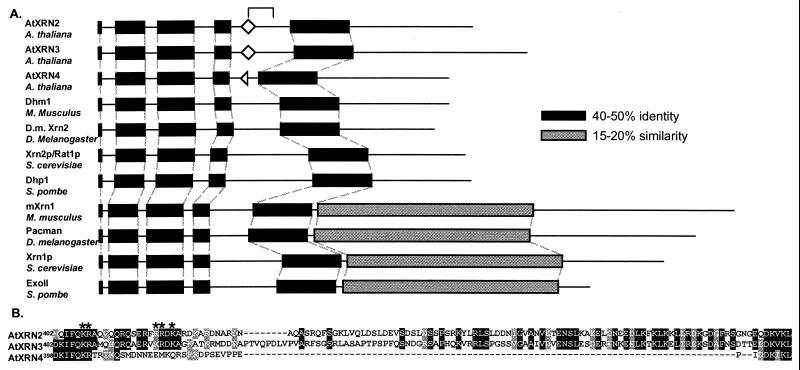
Figure 1.
The Arabidopsis proteins AtXRN2, AtXRN3, and AtXRN4 are orthologs of the Xrn2p/Rat1p protein of S. cerevisiae. (A) Members of the XRN family were aligned with the program clustalw (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html), revealing conserved regions of the XRN family (black regions) and an Xrn1p-specific domain (gray regions). Bipartite NLS consensus motifs (diamonds), in the regions indicated by the bracket, were identified with the program psort (http://psort.nibb.ac.jp). The half diamond in AtXRN4 indicates the lack of the C-terminal portion of a consensus bipartite, NLS. The AtXRNs, M. musculus Dhm1 (accession no. I49635), S. pombe Dhp1 (accession no. S43891), S. cerevisiae Xrn2p/Rat1p (accession no. NP 014691), D. melanogaster gene product (accession AAF52452), Mus musculus mXrn1 (accession no. CAA62820), D. melanogaster Pacman (accession no. CAB43711), S. cerevisiae Xrn1p (accession no. P22147), and S. pombe ExoII (accession no. P40383) were aligned. (B) The amino acid sequences of the AtXRNs, which correspond to the bracket in A, are shown. Identical residues are shown in black; similar residues are in gray. The basic residues constituting a bipartite NLS found in AtXRN2 and AtXRN3, only part of which is conserved in AtXRN4, are indicated by asterisks.