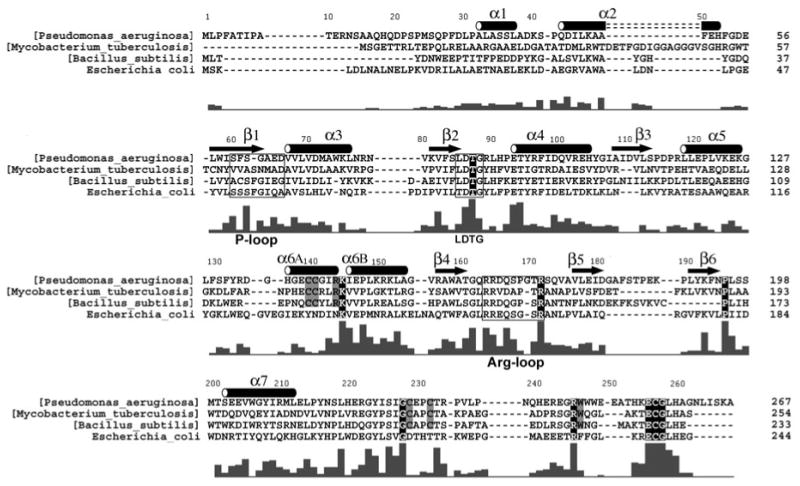
Figure 3.
Primary sequences for APS reductases from P. aeruginosa, M. tuberculosis, and B. subtilis, and for PAPS reductase from E. coli, are shown based on alignment of 38 APS and 34 PAPS reductases (Supplementary Figure 2). Secondary structure elements are denoted for P. aeruginosa APS reductase, and residues within the P-loop (60–66), LDTG motif (85–88), and Arg-loop (163–171), are boxed. The bar graph indicates the degree of conservation based on all 72 sequences (Supplementary Figure 2). Strictly conserved residues are outlined in black; six additional residues conserved in 38 APS reductases that ligate or interact with the [4Fe-4S] cluster are boxed in gray.