Figure 9.

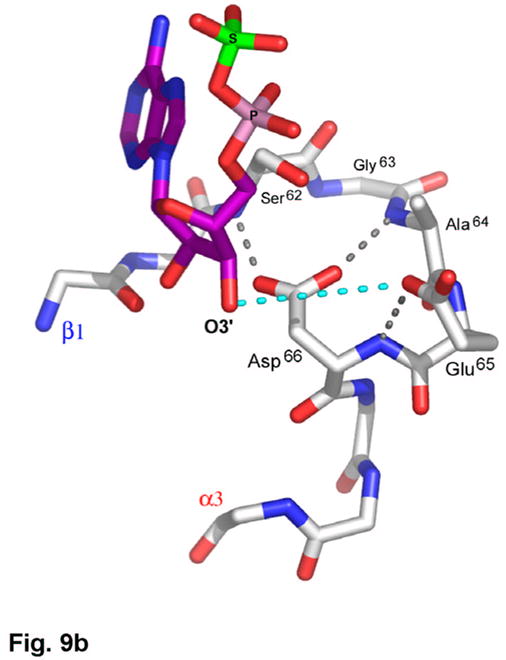
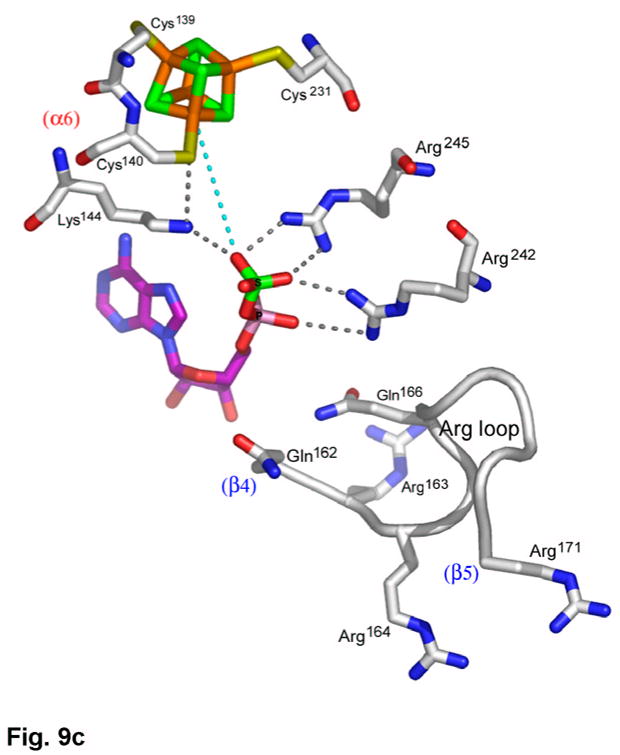
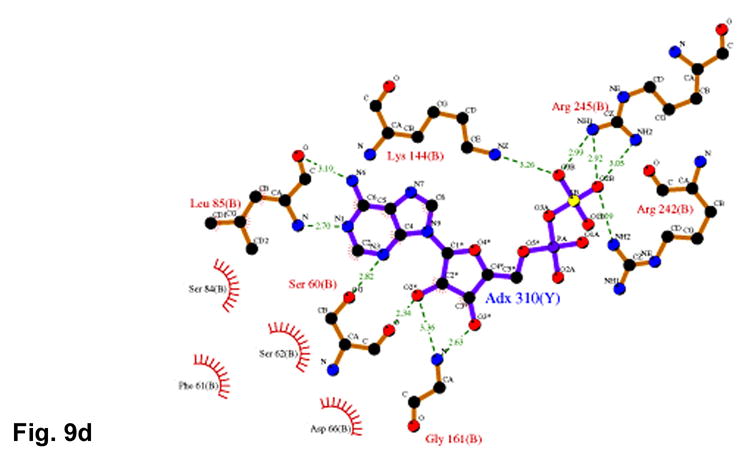
(a) Recognition of the adenosine moiety of APS by conserved residues on strands β1, β2, and β4, which participate in four main chain hydrogen bonds with adenine and the ribose O2′ hydroxyl group. In addition, these residues stabilize the conformation of the β-strands through hydrogen bonds within two conserved motifs (Leu85Asp86Thr87Gly88 and Thr160Gly161). (b) The P-loop comprising conserved residues 60–66 connects strand β1 and helix α3 in the P. aeruginosa APS reductase active site. Three amides hydrogen bond with Glu65 or Asp66, but in PAPS reductases these acidic residues are replaced by Gln and Ala or Ser, respectively. The distance between Glu65 and the O3′ of ribose is 5.3 Å (cyan dotted line). (c) Conserved basic residues in the vicinity of the active site interact with the phosphate and sulfate groups of APS, or reside on the Arg-loop, comprising residues 162–175 between strands β4 and β5. The shortest distance between a sulfate oxygen atom and Fe of the [4Fe-4S] cluster is ~7.0 Å (cyan dotted line). (d) Summary of all active site contacts to APS in subunit B of the asymmetric unit (PDB deposition 2GOY) plotted in two dimensions; hydrogen bond distances are indicated in Å.
