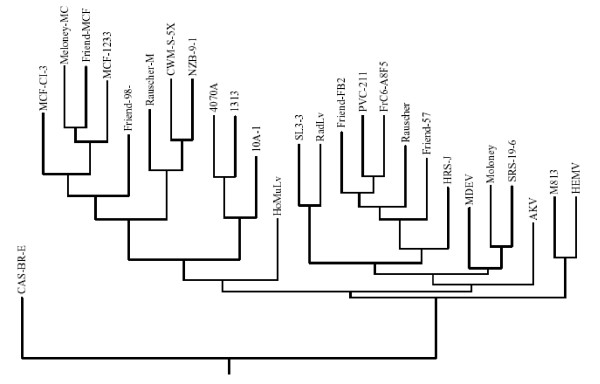
Figure 7.
Phylogenetic analyses of amino acid sequences of complete MuLV env genes. The env genes of 26 MuLV strains were compared with those of MuLV 1313(AF411814) and the evolutionary tree was constructed using deduced amino acid sequences of full-length MuLV env genes and gaps were stripped from the alignment prior to construction of the final tree. Viruses included are: Friend FrC6-A8F5 MuLV (D88386); Friend PVC-211 MuLV (M93134); Rauscher MuLV (U94692); Friend SFFV-spleen focus-forming virus (AF030173); Friend MuLV (X02794); SRS 19-6 MuLV (AF019230) [82]; Moloney MuLV (J02255) [85]; AKV MuLV(J01998) ; Cas-Br-E MuLV (X57540) ; HoMuLV-Mus hortalanus murine leukemia virus (M26527) ; 4070A MuLV (M33469) ; 1313 MuLV(AF411814); 10A1 MuLV (M33470); NZB-9-1 MuLV (K02730); CWM-S-5X MuLV (M59793); HRS/J MuLV (M17326); Friend 98 MCF-mink cell focus-forming virus (AF133256); Friend MCF (X01679); Rauscher MCF (M10100); Moloney MCF (J02254); CI-3 MCF (K02725), 1233 MCF (U13766), MDEV (AF053745), HEMV (AY818896), M813 (AF327437) RadLV (K03363) and SL3-3 (AF16925). Note the unique separation of amphotropic viruses on a single node with a distinct branch for MuLV-1313.