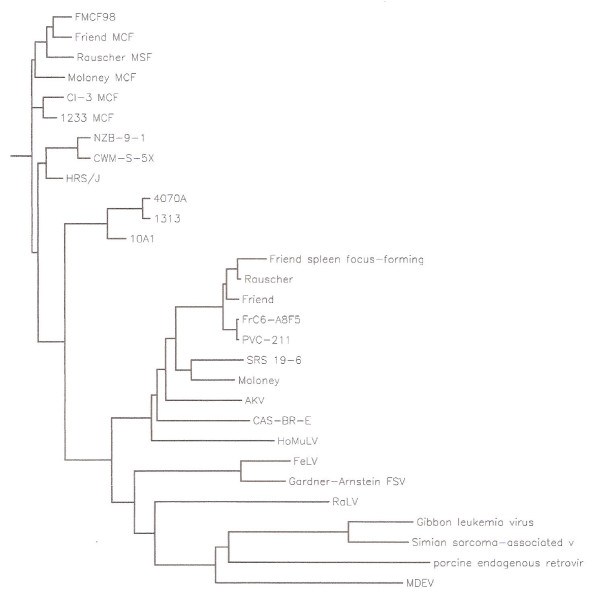
Figure 8.
Phylogenetic relationship of MuLV 1313 to various mammalian gammaretroviruses. This analysis utilized full-length or nearly full-length Env proteins 29 retroviruses isolated from mouse, rat, cat, pig, monkey and gibbon ape. The original multiple sequence alignment was created with Clustal W and the rooted tree phenogram was constructed using pre-aligned sequences and PHYLIP [64-66, 71, 80]. Abbreviations and accession numbers (in brackets) of viruses used in the phenogram are according to the GenBank database SSAV-simian sarcoma-associated virus (AF055064); GALV-gibbon ape leukemia virus (M26927); PERV-porcine endogenous virus (Y17013); MDEV-Mus dunni endogenous virus (AF053745); RaLV-rat leukemia virus (M77194), FeLV A-feline leukemia virus, subgroup A (M18247); FeLV B-feline leukemia virus, subgroup B (Gardner-Arnstein strain) (X00188); MuLV-murine leukemia virus MuLV 1313 (AF411814); Friend FrC6-A8F5 MuLV (D88386); Friend PVC-211 MuLV (M93134); Rauscher MuLV (U94692); Friend SFFV-spleen focus-forming virus (AF030173); Friend MuLV (X02794); SRS 19-6 MuLV (AF019230) [83]; Moloney MuLV (J02255); AKV MuLV(J01998) ; Cas-Br-E MuLV (X57540) ; HoMuLV-Mus hortalanus murine leukemia virus (M26527) ; 4070A MuLV (M33469) ; 10A1 MuLV (M33470); NZB-9-1 MuLV (K02730); CWM-S-5X MuLV (M59793); HRS/J MuLV (M17326); Friend 98 MCF-mink cell focus-forming virus (AF133256); Friend MCF (X01679); Moloney MCF (J02254); CI-3 MCF (K02725), 1233 MCF (U13766), MDEV (AF053745), RadLV (K03363) and SL3-3 (AF16925)