Abstract
Background
The quail and chicken major histocompatibility complex (Mhc) genomic regions have a similar overall organization but differ markedly in that the quail has an expanded number of duplicated class I, class IIB, natural killer (NK)-receptor-like, lectin-like and BG genes. Therefore, the elucidation of genetic factors that contribute to the greater Mhc diversity in the quail would help to establish it as a model experimental animal in the investigation of avian Mhc associated diseases.
Aims and approaches
The main aim here was to characterize the genetic and genomic features of the transcribed major quail MhcIIB (CojaIIB) region that is located between the Tapasin and BRD2 genes, and to compare our findings to the available information for the chicken MhcIIB (BLB). We used four approaches in the study of the quail MhcIIB region, (1) haplotype analyses with polymorphic loci, (2) cloning and sequencing of the RT-PCR CojaIIB products from individuals with different haplotypes, (3) genomic sequencing of the CojaIIB region from the individuals with the different haplotypes, and (4) phylogenetic and duplication analysis to explain the variability of the region between the quail and the chicken.
Results
Our results show that the Tapasin-BRD2 segment of the quail Mhc is highly variable in length and in gene transcription intensity and content. Haplotypic sequences were found to vary in length between 4 to 11 kb. Tapasin-BRD2 segments contain one or two major transcribed CojaIIBs that were probably generated by segmental duplications involving c-type lectin-like genes and NK receptor-like genes, gene fusions between two CojaIIBs and transpositions between the major and minor CojaIIB segments. The relative evolutionary speed for generating the MhcIIBs genomic structures from the ancestral BLB2 was estimated to be two times faster in the quail than in the chicken after their separation from a common ancestor. Four types of genomic rearrangement elements (GRE), composed of simple tandem repeats (STR), were identified in the MhcIIB genomic segment located between the Tapasin-BRD2 genes. The GREs have many more STR numbers in the quail than in the chicken that displays strong linkage disequilibrium.
Conclusion
This study suggests that the Mhc classIIB region has a flexible genomic structure generated by rearrangement elements and rapid SNP accumulation probably as a consequence of the quail adapting to environmental conditions and pathogens during its migratory history after its divergence from the chicken.
Background
The genomic region of the major histocompatibility complex (Mhc) contains multi-gene family members involved in the immune response. The Mhc class I and class II genes encode glycoproteins that transport foreign peptides to the surface of cells for recognition by T cell receptors on lymphocytes, which in turn kill infected cells [1]. The Mhc class II molecules have highly polymorphic peptide binding regions (PBR) for the α1 and β1 domains encoded by the class IIA and class IIB genes, respectively, in various vertebrates including avian [2]. These polymorphisms may have been generated by gene conversion and positive selection, such as balancing selection and overdominant selection to adapt to life-environmental pathogens [3,4].
The most information currently available on the genomic organization of the Mhc in birds is for the chicken and quail. The chicken (Gallus domesticus) Mhc (Gado) region is divided into two major parts, Gado-B and Gado-Y [5]. Both of these regions are inherited independently of each other, although they are physically linked on micro-chromosome 16 (GGA16), [6-9]. From previous genomic studies, GGA16 is suggested to be an essential immunity chromosome, which is composed of genes involved with adaptive immunity (BF/BL segment in Gado-B), innate immunity (Gado-Y) and intrinsic immunity (TRIM-like and BG gene segments in the extended Gado-B region [[5,10-13], Shiina et al, Unpublished data].
The quail (Coturnix japonica, Coja) belongs to the same order (Galliformes) and family (Phasianidae) as the chicken. Whereas the quail is a migratory bird originating from Northern and Southern Asia, flying only short distances at a time and with a relatively short history of domestication, the chicken is a non-migratory bird originating from Southeastern Asia and with a thousand year history of domestication. Chicken – quail hybrids have been produced by artificial insemination of quails with chicken sperm [22]. Several immunological traits have been compared between various lines of quail which were selected for high and low IgY levels in the serum [23], and for high and low secondary antibody responses to Newcastle disease virus [24,25], influenza virus, sheep erythrocytes and Salmonella pullorum [26]. The quail is susceptible to Marek's disease virus as observed in the chicken. The chicken Gado-B complex has a significant influence in the genetics of disease resistance, such as Rous sarcoma and Marek's disease, but which genes within the complex are responsible and how they confer resistance or susceptibility is not known [16-18]. One important obstacle to elucidating the Mhc resistance genes in chicken is that the BF, BL and BG genes have co-evolved as haplotypes with a high linkage disequilibrium between the genes due to the compact structure of its "minimal essential" Mhc [19-21]. However, the immune response against the Marek's disease tumor-associated surface antigen (MATSA) in the quail differs significantly from that in the chicken [27]. Such immunological differences are likely to be due in part to the number and variation in Mhc gene loci and/or alleles, but the data concerning the Coja haplotypes and their association with disease is largely lacking. From our previous transcription and genomic studies, we found that the quail and chicken Mhc regions have a similar overall organization, but differ markedly in that the quail has an expanded number of duplicated genes with 7 class I, 10 class IIB, 4 natural killer (NKr)-like receptor, 6 lectin-like receptor and 8 BG genes [14,28,29]. To explain these findings, haplotypic genome comparison among quails and between the quail and the chicken is a matter of primary importance to elucidate the gene organization, molecular mechanism of polymorphism generation and disease analysis.
In order to better understand the genetic factors involved in the generation of Mhc diversity in quail, we identified and characterized six CojaIIB haplotypes by genotyping and polymorphism analysis using expressed CojaIIB sequences. We determined the genomic sequences within the Tapasin-BRD2 genomic segment of five different Coja haplotypes including the major transcribed CojaIIBs. We also compared the quail Mhc class II genomic structures and diversities of the chicken B12 and B21 haplotypic orthologs by phylogenetic analysis and duplication modeling. This study shows that the quail has much greater Mhc class IIB diversity and genomic structural flexibility than the chicken even though the two species are closely related in the evolutionary spectrum.
Results and discussion
Coja haplotype analysis with polymorphic loci
Three polymorphic markers PM1, PM2 and PM3, (Table 1) were used to genotype 48 randomly selected quails. We chose these three markers for the preliminary classification of Coja haplotypes because they are located in the Tapasin, Coja-DBB1 and Coja-DMB2 loci, respectively (Figure 1). The allele frequencies and heterozygosities for the polymorphic markers are presented in Table 2. All of the genotype frequencies were over 13% and the heterozygosity for each marker was over 0.65. The minimum number of haplotypes predicted from the maximum likelihood analysis of the 48 quails by the two different methods, a Bayesian statistical method and an Expectation-Maximization (EM) algorithm, was six (Table 3). Of the six haplotypes, the haplotype numbers (HT1 to HT5 had higher frequencies, ranging from of 14.6% to 28.1%, than HT6 (5.2%). Interestingly, HT2 and HT6 had the same "6–10" and "*02" allele combinations at the PM1 and PM2 loci respectively, but a different allele type at the PM3 locus (Table 3).
Table 1.
Basic features of the three polymorphic markers on Coja region
| Coja haplotype 1 (Acc. Num. AB078884) | |||||||
| marker name | Nuc. position** | Gene position | length (bp) | range (bp) | polymorphisms | primer name | primer sequence (5' to 3') |
| PM1 | 89043 – 89887 | Tapasin (ex.4 – int.4) | 845 | 718–1095 | (ATGAT)n (TTCCTATGGGGGCTGTAGGGTGGATGGGACTGGGTGGTA)n |
TAPBPL-F | CAGGTCCTGCTGGCCTATGA |
| TAPBPL-R | TGGTGTGATGCCAACCCAT | ||||||
| PM2 | 92768 – 93787 | Coja-DBB1 (Promoter – ex.3) | 1002 | 989–1002 | 126 SNPs, 13 indels | CojaDBBpromoterF6 | CCCTGGGGACACCATTTGTCAT |
| C2BNO5 | GCGCCAGGAAGACGAGCCCCAGCAC | ||||||
| PM3* | 118204 – 118536 | DMB2 (int.1 – int.2) | 333 | 333 | G/A, G/C | DMB2-F | GGAGTGCATCCCCATTGCT |
| DMB2-R | GCTCACTCTTGCGCAGTGC | ||||||
* PM3 contains two SNP sites.
** positions on Coja haplotype 1 genomic sequence (accession number; AB078884).
Figure 1.
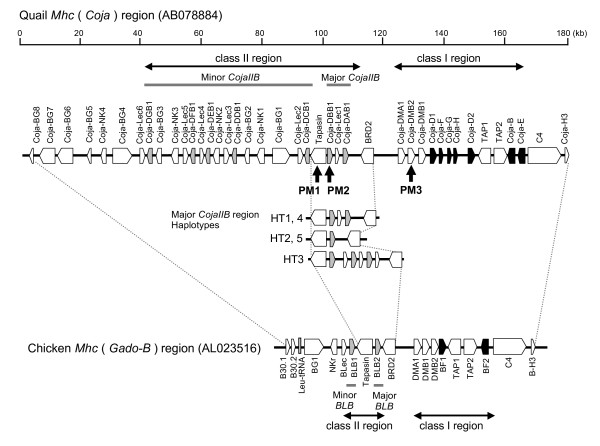
Comparative gene map of quail and chicken Mhc region and locations of the genetic markers PM1 to PM3. The map shows the comparison of the 180 kb Coja haplotype 1 sequence (AB078884: Shiina et al 2004) and the 92 kb chicken B12 haplotype sequence (AL023516: Kaufman et al 1999). Black and gray boxes indicate the Mhc class I and class IIB loci respectively. White boxes indicate other genes. The labeled vertical arrows indicate the locations of the newly designated genetic markers PM1 – PM3.
Table 2.
Genotype frequency and heterozygosity for three markers
| marker | Allele | length (bp) | Nt. Acc, No. | Frequency (%) | Heterozygosity |
| PM1 | 9–12 | 845 | AB282647 | 28.1 | 0.794 |
| 6–10 | 718 | AB282648 | 22.9 | ||
| 8–12 | 806 | AB282649 | 13.5 | ||
| 10–14 | 900 | AB282650 | 20.8 | ||
| 18–14 | 1095 | AB282651 | 14.6 | ||
| PM2 | *01 | 1002 | AB282652 | 28.1 | 0.794 |
| *02 | 1001 | AB282653 | 22.9 | ||
| *03 | 989 | AB282654 | 13.5 | ||
| *04 | 1001 | AB282655 | 20.8 | ||
| *05 | 989 | AB282656 | 14.6 | ||
| PM3 | G-C | 333 | AB186377 | 28.1 | 0.650 |
| G-G | 333 | AB186378 | 45.8 | ||
| A-C | 333 | AB186379 | 26.0 |
Table 3.
Summary of Coja haplotypes (HT1–6) and their frequencies
| HT num. | PM1 | PM2 | PM3 | Obs. Num. | Freq. (%) |
| 1 | 9–12 | *01 | G-C | 27 | 28.1 |
| 2 | 6–10 | *02 | G-G | 17 | 17.7 |
| 3 | 8–12 | *03 | G-G | 13 | 13.5 |
| 4 | 10–14 | *04 | A-C | 20 | 20.8 |
| 5 | 18–14 | *05 | G-G | 14 | 14.6 |
| 6 | 6–10 | *02 | A-C | 5 | 5.2 |
Bold letter shows Coja haplotype frequency with over 10%.
On the basis of finding the different allele type at the PM3 locus in HT6, recombination happened at least once in the region within the 24.4 kb segment between the quail genes Coja-DBB1 and Coja-DMB2 (PM2–PM3) and the newly created haplotype has spread in the population. Future analyses using fully pedigreed families should help to ascertain whether the intervening segments of DNA are the same in different individuals with the same three-marker genotype. Nevertheless, the variability in the quail genomic segment between the Coja-DBB1 and Coja-DMB2 genes corresponds to the chicken BF/BL region that displays strong linkage disequilibrium and genomic structural stability [19,20].
Gene loci identification and haplotype reanalysis by RT-PCR, cDNA cloning and sequencing of transcribed CojaIIB loci
In order to identify and characterize the transcript sequences expressed by the gene loci of the different haplotypes, six quails with relatively high HT frequencies representing haplotypes HT1 to HT5 were selected for RT-PCR, cDNA cloning and sequencing analysis. The transcription intensity of the CojaIIB genes was estimated from the number of clones sequenced. A summary of the transcription intensity results for the five haplotypes is presented in Table 4. The Table shows the transcription intensity for HT1 to HT5 as a percent frequency of the clones for each individual quail (identified as numbers 302, 311, 312, 321, 322 and 323) with the percent frequency of clones per haplotype in parenthesis. The Table also shows the gene locus, the detected allele type, the PM2 allele, the nucleotide accession number in GenBank and the percent nucleotide similarity of the cDNA clones to the transcribed sequences expressed by the CojaIIB loci of HT1.
Table 4.
Percent frequency of cloned cDNA, distribution and classification of the transcribed CojaIIB allele as five distinct Coja haplotypes
| HT number | Locus | Allele | PM2 alleles | Nt. Acc, No. | Nucleotide similarity to HT1 (%) | % frequency of cDNA clones per individual quail (% frequency of cDNA clones per haplotype) | |||||
| quail individual number | |||||||||||
| 302* | 311 | 312 | 321 | 322 | 323 | ||||||
| HT1 | Coja-DAB1 | Coja-DAB1*01 | AB181861 | 100 | 46.9** (46.9)*** | - | 17.1 (37.1) | 5.9 (19.0) | - | - | |
| Coja-DBB1 | Coja-DBB1*01 | Coja-DBB1*01 | AB181862 | 100 | 35.9 (35.9) | - | 22.4 (48.6) | 16.2 (52.4) | - | - | |
| Coja-DCB1 | Coja-DCB1*01 | AB181863 | 100 | 6.3 (6.3) | - | 1.3 (2.9) | 2.9 (9.5) | - | - | ||
| Coja-DDB1 | - | - | - | - | - | - | - | - | - | ||
| Coja-DEB1 | Coja-DEB1*01 | AB181865 | 100 | 4.7 (4.7) | - | 2.6 (5.6) | - | - | - | ||
| Coja-DFB1 | Coja-DFB1*01 | AB181866 | 100 | - | - | 1.3 (2.9) | 1.5 (4.8) | - | - | ||
| Coja-DGB1 | Coja-DGB1*01 | AB181868 | 100.0 | 6.3 (6.3) | - | - | 4.4 (14.3) | - | - | ||
| Coja-DGB1 | Coja-DGB1*02 | AB181869 | 98.9 (DGB1) | - | - | 1.3 (2.9) | - | - | - | ||
| HT2 | CojaII-13 | CojaII-13*01 | Coja-DBB1*02 | AB181874 | 76.3 (DCB1) ~ 87.4 (DAB1) | - | 62.9 (100) | 53.9 (100) | - | - | 67.2 (100) |
| HT3 | CojaII-16 | CojaII-16*01 | Coja-DBB1*03 | AB264281 | 78.9 (DCB1) ~ 85.2 (DBB1) | - | 12.9(34.8) | - | - | - | - |
| CojaII-17 | CojaII-17*01 | AB264282 | 77.5 (DCB1) ~ 83.3 (DGB1) | - | 24.2 (65.2) | - | - | - | - | ||
| HT4 | CojaII-02 | CojaII-02*01 | Coja-DBB1*04 | AB181871 | 78.1 (DCB1) ~ 88.9 (DBB1) | - | - | - | 13.2 (19.1) | 14.5 (29.4) | - |
| CojaII-01 | CojaII-01*01 | AB181870 | 78.9 (DCB1) ~ 87.8 (DBB1) | - | - | - | 36.8 (53.2) | 21.7 (44.1) | - | ||
| CojaII-04 | CojaII-04*01 | AB181872 | 77.8 (DCB1) ~ 84.4 (DBB1) | - | - | - | 19.1 (27.7) | 13.0 (26.5) | - | ||
| HT5 | CojaII-14 | CojaII-14*01 | Coja-DBB1*05 | AB181876 | 77.4 (DCB1) ~ 87.4 (DAB1) | - | - | - | - | 47.8 (94.3) | 28.1 (85.7) |
| Coja-DFB1 | Coja-DFB1*02 | AB181867 | 99.3 (DFB1) | - | - | - | - | 2.9 (5.7) | 4.7 (14.3) | ||
| cDNA sub-clone | 64 | 62 | 76 | 68 | 69 | 64 | |||||
| Presumptive HT | 1/1 | 2/3 | 1/2 | 1/4 | 4/5 | 2/5 | |||||
Bold letter indicates sub-clone frequency over 30%. (-) means not transcribed. Coja-DFB1*02 was categorized to "HT5" because this allele sequence was observed in only HT5.
In total, 15 kinds of CojaIIB cDNA sequences derived from the RT-PCR products of six quails were identified by sequence comparison (Table 4). Another 19 CojaIIB sequences were previously reported [14,25]. Altogether, these expressed sequences were identified to be expressed either by loci that were previously described alphabetically as Coja-DAB1 to Coja-DGB1 for HT1 or by as yet unnamed loci. The new loci identified in this study were therefore given names, such as CojaII-01, CojaII-02, CojaII-13 and CojaII-16 as listed in Table 4, using the naming formulae "CojaII-XX" as suggested by Shimizu et al [25] for the nomenclature of CojaIIB sequences. Each quail transcribed three to eight kinds of CojaIIB gene sequences (Table 4). Five to six of the cDNA sequences, which were observed in three quails (302, 312 and 321), perfectly matched with six of the seven CojaIIB loci of Coja haplotype 1 (HT1), that is Coja-DAB1 and -DBB1 in the major class II region, and Coja-DCB1, -DEB1, -DFB1 and -DGB1 in the minor class II region [14]. However, no sequences were detected in our study for the HT1 Coja-DDB1 locus. In addition, two other cDNA sequences were tentatively named Coja-DFB1*02 in HT5 and Coja-DGB1*02 in HT1 because they appear to be additional alleles at the Coja-DFB1 and -DGB1 loci, respectively, as determined from our phylogenetic analysis (Figs. 2, Table 4, Additional file 1). The sequences of CojaII-01, -02 and -04 were perfectly matched to the previously determined CojaII-01HL, -02HL and -04H haplotype sequences (Accession numbers; AB110476 to AB110478), respectively, which were derived from different inbred lines [25,26]. However, the CojaII-13, -14, -16 and -17 sequences did not have significantly high nucleotide similarities with the previously determined sequences (76.3 ~ 88.9%) of HT1 sequences and, therefore, were assigned as belonging to other unique haplotypes (Table 4).
Figure 2.
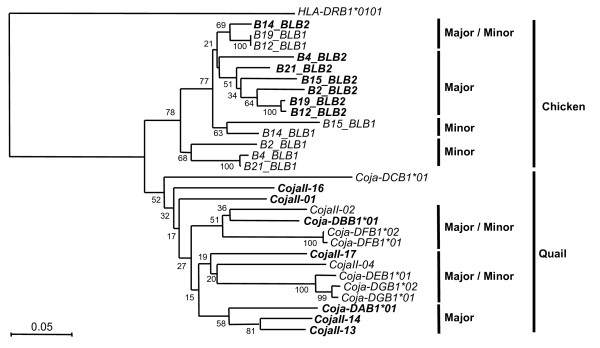
Phylogenetic tree of the quail and chicken transcribed MhcIIBs. The dendrogram was constructed from the nucleotide sequences of the β1 extracellular domain regions (exon 2) (270 nucleotides in length) of MhcIIB genes for the 16 CojaIIBs shown in figure S1 the chicken haplotypic BLB sequences on B2 B4 B12 B14 B15 B19 and B21 (AJ248576 AJ248583 AJ248572 AJ248581 AJ248585 AJ248577 AJ248584 AJ248579 AJ248580 AJ248582 AJ248574 AJ248586 AJ248575 and AJ248573) and HLA-DRB1 (AF142457). The HLA-DRB1 sequence used as a species outlier roots the tree. Values near the branch-points of the tree indicate the bootstrap values. Bold letters indicate major transcribed CojaIIBs and BLBs.
On the basis of the correlation of the Coja haplotypes and distribution of the transcribed CojaIIBs for each of the quails, the fifteen CojaIIB sequences were classified to five distinct Coja haplotypes, namely, HT1, consisting of Coja-DAB1, -DBB1, -DCB1, -DEB1, -DFB1 and -DGB1 loci; HT2, CojaII-13; HT3, CojaII-16 and -17; HT4, CojaII-01, -02 and -04; and HT5, Coja-DFB1 and CojaII-14 (Table 4). From the cDNA cloned frequencies per Coja haplotype, Coja-DAB1, -DBB1, CojaII-01, -13, -14, -16 and -17, excluding -DAB1 in quail 321, were the major transcribed CojaIIBs with cloned frequencies of 34.8 ~ 100%, CojaII-02 and -04 were moderately transcribed CojaIIBs with cloned frequencies of 19.1 ~ 29.4%, and Coja-DCB1*01, -DEB1*02, -DFB1*01, -DFB1*02, -DGB1*01 and -DGB1*02 were minor transcribed CojaIIBs with cloned frequencies of 2.9 ~ 14.3% (Table 4). This result suggests that each Coja haplotype has at least one and up to seven transcribed CojaIIB loci with one or two of them representing the major locus.
Genomic diversity of Tapasin-BRD2 segment
The Tapasin-BRD2 genomic segment contains the major CojaIIB region with the Coja-DAB1, Coja-DBB1 and the Coja-Lec1 genes flanked by the Tapasin and the BRD2 genes. This segment also has the PM1 and PM2 markers that we used for haplotyping. In order to study the genomic diversity of the Tapasin-BRD2 segments in different haplotypes by genomic sequencing, two cosmid libraries were constructed representing the haplotypes HT2, HT3, HT4 and HT5.
The average insert sizes of cosmid libraries constructed from the genomic DNA of quails 311 (HT 2/3) and 322 (HT 4/5) were estimated to average 37.5 kb and 44.6 kb, respectively, by 0.3% agarose gel electorophoresis analysis using 20 randomly selected cosmids (data not shown). As the quail has a genome size of 1.2 × 109 bp, similar to the chicken, the two cosmid libraries 311 and 322 were expected to cover 2.5 × 1010 bp (20.9-fold) and 21.4 × 1010 bp (17.8-fold), respectively [11]. Two Tapasin (clone number: 311CIIB-18 and 311CIIB-20) and two BRD2 positive cosmids (322CIIB-01 and 322CIIB-02) were isolated from HTs 2/3 and HTs 4/5 cosmid libraries, respectively. DNA typing revealed that the CojaIIB genes within the cosmids 311CIIB-18, 311CIIB-20, 322CIIB-01 and 322CIIB-02 corresponded to the haplotypes HT2, HT3, HT5 and HT5, respectively. Therefore, we selected the cosmids 311CIIB-18 (HT2), 311CIIB-20 (HT3) and 322CIIB-02 (HT5) for the genomic sequencing study of the Tapasin-BRD2 segment.
The genomic sequences derived from 311CIIB-18, 311CIIB-20 and 322CIIB-02 cosmids were determined by the shotgun method. Also a part of the Tapasin-BRD2 segment was sequenced from the long-ranged PCR products amplified from the genomic DNA of HT3 and HT4 homozygote quails [25,26]. The genomic structures of the major transcribed MhcIIB segments within the 5 quail haplotypes H1 to H5 and the chicken haplotype B12 derived from the genomic sequencing information and transcript alignments are shown in Fig. 3. The dot-matrix analyses between the 5 Coja haplotypes, chicken and quail, and two chicken haplotypes are shown in Additional file 2. After sequencing, the nucleotide lengths of the segment between the Tapasin-BRD2 genes were determined to be 5,984 bp in HT2, 16,462 bp in HT3, 5,222 bp in HT4 and 5,840 bp in HT5. The sequence of four segments on HT3 and HT4 was not determined fully however, for DNA structural reasons, such as the difficulties encountered with long repeat sequences and extremely high GC contents (Figs. 3, Additional file 2).
Figure 3.
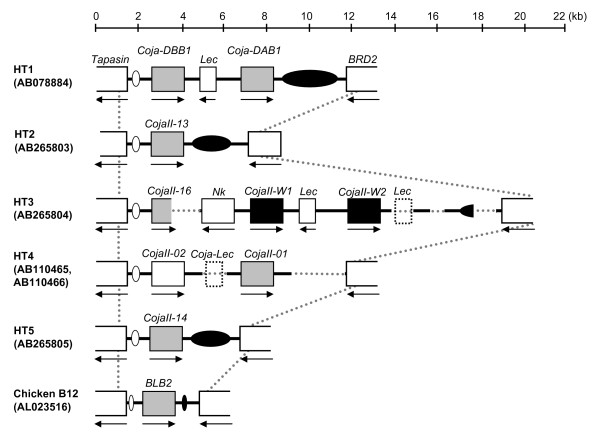
Genomic structures of the major transcribed MhcIIB segments in the quail and chicken. The map shows the comparison of the major transcribed MhcIIB segments on five Coja haplotypic sequences (HT1 HT2 HT3 HT4 and HT5) and the chicken B12 haplotype sequence. Gray striped and black boxes indicate the major transcribed MhcIIB moderately transcribed MhcIIB and non-transcribed MhcIIB respectively. White boxes indicate other genes. The numbers in parenthesis are the GenBank accession numbers. The white oval indicates TB1 and the black oval indicates the location of TB2-1 TB2-2 and TB3.
The Tapasin-BRD2 segment showed extremely complicated genome structures among the Coja haplotypes examined, similar to the HLA-DR region in the human MHC genomic region (Figs. 3, Additional file 1) [31]. The Tapasin-BRD2 segment of HT2 and HT5 contained only one major transcribed CojaIIB (CojaII-13 and -14, respectively) and their genomic structures and nucleotide length were similar to the chicken orthologous BL region (Figs. 3, Additional file 1). The Tapasin-BRD2 segment of HT1 contained two major transcribed CojaIIBs (Coja-DAB1 and -DBB1) (Fig. 3) [13]. Similarly, the Tapasin-BRD2 segment of the HT4 contained two CojaIIBs (CojaII-01 and CojaII-02) with major and moderately transcribed CojaIIB genes (Table 4). In HT3, the Tapasin-BRD2 segment contained three CojaIIBs, CojaII-16, Coja-W1 and Coja-W2. The CojaII-16 is a major transcribed CojaIIB, but although both Coja-W1 and -W2 have intact structures their mRNA was not obtained from the peripheral blood cells (Fig. 3). In all of the major transcribed CojaIIBs, the nucleotide length of intron 1 was variable whereas the other introns were relatively well conserved (Table 5). Moreover, one major transcribed CojaIIB (CojaII-17) that was observed in the HT3 (Table 4) was not identified in the Tapasin-BRD2 segment (Additional file 1). Consequently, the CojaII-17 is the only gene that either locates to a minor transcribed CojaIIB locus or to some other genomic region.
Table 5.
Comparison of nucleotide length of intron and exons among CojaIIB in the Tapasin-BRD2 segment
| Coja HT | HT1 | HT2 | HT3 | HT4 | HT5 | ||
| CojaIIB PM2 alleles | Coja-DAB1 | Coja-DBB1 Coja-DBB1*01 | CojaII-13 Coja-DBB1*02 | CojaII-16 Coja-DBB1*03 | CojaII-01 | CojaII-02 Coja-DBB1*04 | CojaII-14 Coja-DBB1*05 |
| exon1 | 91 | 91 | 91 | 91 | 91 | 91 | 91 |
| intron1 | 30 | 88 | 88 | 76 | 146 | 88 | 76 |
| exon2 | 270 | 270 | 270 | 270 | 270 | 270 | 270 |
| intron2 | 86 | 86 | 86 | 86 | 86 | 86 | 86 |
| exon3 | 282 | 282 | 282 | 282 | 282 | 282 | 282 |
| intron3 | 95 | 95 | 95 | 95 | 95 | 95 | 95 |
| exon4 | 111 | 111 | 111 | 111 | 111 | 111 | 111 |
| intron4 | 76 | 76 | 76 | 76 | 76 | 77 | 76 |
| exon5 | 24 | 24 | 24 | 24 | 24 | 24 | 24 |
| intron5 | 73 | 86 | 73 | 73 | 73 | 73 | 73 |
| exon6 | 14 | 14 | 14 | 14 | 14 | 14 | 14 |
| total | 1152 | 1223 | 1210 | 1198 | 1268 | 1211 | 1198 |
In summary, the Tapasin-BRD2 segment contains one or two major transcribed CojaIIBs that were identified in this study in much the same way as the chicken with its major transcribed MhcIIB, BLB2 (Figs. 1, 3). In contrast to Xenopus and mammals that also have MhcIIB genes located within the Tapasin-BRD2 segment, the bony fishes, such as medaka, fugu and rainbow trout, have MhcI genes within the Tapasin-BRD2 segment. Thus, the locations of the major transcribed CojaIIBs and BLB2 are well conserved in birds and more comparable with mammals and reptiles than with bony fish from the point of view of evolution [15,32-34].
Molecular evolutionary analysis of the chicken and quail MhcIIB genomic region
In a comparison of the CojaIIB gene organization of different haplotypes, the CojaIIB genomic units in the Tapasin-BRD2 segment of the HT1 [DBB1 – Lec – DAB1] and HT4 [CojaII-02 – Lec – CojaII-01] were noted to resemble the minor CojaIIB genomic DFB1/DEB1 unit [DFB1 – Lec4 – DEB1], whereas those of the HT3 [CojaII-W2 – Lec – CojaII-W1 – Nkr – CojaII-16] resemble the DFB1/DEB1/DDB1 unit [DFB1 – Lec4 – DEB1 – NK2 – (Lec3) – DDB1], although they are in the opposite direction to each other (Fig. 3). Thus, two kinds of successive trans-segmental duplications involving non-Mhc genes appear to have produced the major CojaIIB segments independently. However, significant nucleotide homologies between the duplicated units were not observed (data not shown). Since comparable segmental duplication variability has not been observed in the Gado-B region, the trans-segmental duplications in quail are likely to have occurred after speciation of the quail and chicken from their common ancestor. In the case of human MHC, the traits of segmental duplications and transpositions were observed in the HLA class I region [35], and especially the generation of the HLA-B and -C segment that is explained by the events involving the MHCI, MIC, HCGII, HCGIV, HCGIX, HCP5, 3.8-1 unit in the evolutionary process [36].
To clarify the genetic relationships among the CojaIIB sequences and between the quail and the chicken MhcIIB sequences, a phylogenetic tree of exon 2 was constructed (Fig. 2). This tree suggests that the CojaIIB gene sequences are more closely related to each other in the quail than to the chicken MhcIIB (BLBs), and that the CojaIIB genes have been duplicated after speciation of quail and chicken. The average genetic distance of the CojaIIB sequences (0.194+0.018) after speciation is twice as long as that of the BLB sequences (0.107+0.016), meaning that the CojaIIB genes have a relatively faster evolutionary speed than the BLBs (Fig. 2). On the other hand, chicken haplotype sequences are more closely related. From the genome sequencing of the Red Jungle Fowl Mhc region, the BLB nucleotide sequences were perfectly matched with domestic chicken B21*BLB1 and B21*BLB2 sequences [Shiina et al, Unpublished data]. Therefore, the BLB genes appear to have been generated from a recent common ancestor of both the egg-laying domestic chicken and broilers.
In the phylogenetic tree, the quail lineage was divided into three main clusters, one major and two major/minor intermingled clusters, with the Coja-DCB1*01, CojaII-01 and CojaII-16 out-grouped from the three main clusters. In comparison, the chicken lineage was divided into four main clusters, namely one major, two minors and one major/minor intermingled cluster (Fig. 2). In the case of the major transcribed CojaIIBs, these were observed in all clusters. In addition, the traits for two kinds of segmental duplications were observed in the Tapasin-BRD2 segment as previously mentioned, suggesting that the major transcribed CojaIIBs were generated by independent duplications and/or transpositions between the major and minor CojaIIB segments at a relatively high evolutionary speed after the separation of the quail and chicken from their common ancestor (Fig. 4). In contrast, the BLB2 sequences of extant chickens appear to have been generated from an ancestral BLB2 gene with very little structural reorganization or change (Fig. 4).
Figure 4.
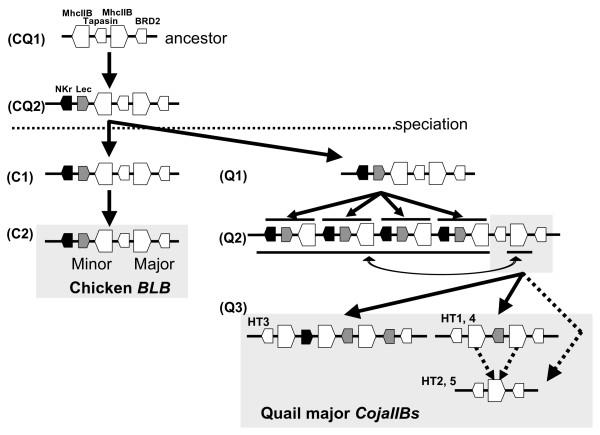
Gene duplication models inferred from the reconstructed phylogenetic trees. The labeled arrows indicate the speciation and gene duplication events for the chicken (C) and quail (Q) lineages. Dotted arrows indicate the evolutionary process for the origin of HT2 and HT5. Gray backgrounds indicate quail and chicken MHCIIB structures at the present-day.
Genomic rearrangement elements in the major CojaIIB segment involved in the generation of Mhc diversity
Four types of tandem repeat sequences were identified that possibly drive genomic rearrangement events within the Tapasin-BRD2 segment of HT1, HT2, HT3 and HT5 (Table 6). These simple candidate rearrangement elements are TB1, a known T-cell factor motif; TB3 and TB3, STRs found in the mouse Mhc class II region; and TB4, a recombination motif [37-39]. From the comparison of the repeat numbers of the solitary repeat unit between quail and chicken, the Coja haplotypes were found to contain numerous repeat numbers for TB1 to TB4, whereas the chicken B12 and B21 haplotypes were found to have none or only a few repeat numbers (Table 6). The TB repeats were not identified in the minor transcribed CojaIIB segment of the HT1 genome contigs [14]. Therefore, the TB repeats appear to be rearrangement elements within the genome that provide strong driving forces for the generation of new major transcribed CojaIIBs via duplications and/or transpositions.
Table 6.
Candidate rearrangement elements (TB1–4) identified within the Tapasin-BRD2 segment
| Name | Features and sequence of repeat unit | Quail | Chicken | |||||
| HT1 | HT2 | HT3 | HT4 | HT5 | B12 | B21 | ||
| TCF-I [37] | ||||||||
| TB1 | MAMAGn | 13* | 13 | 26 | 27 | 23 | 3 | 3 |
| STRs in H-2 class II region [38] | ||||||||
| TB2 | CAGAn | 86 | 54 | >14 | Un | 56 | 5 | 5 |
| TB3 | CAGGn | 120 | 75 | >21 | Un | 99 | 6 | 6 |
| Recombination hotspot motif [39] | ||||||||
| TB4 | GGGCAGGARGn | 9 | 4 | >1 | Un | 5 | 1 | 1 |
| Total | 228 | 146 | >62 | >27 | 183 | 15 | 15 | |
* number of repeat units.
Genomic diversity within the major CojaIIB region and disease studies
Mhc diversity in vertebrates is often attributed to the high SNP content in this genomic region. The generation of nucleotide polymorphism of Mhc genes is usually explained by positive selection, such as balancing selection and overdominant selection acting on the Mhc genes, which is necessary to maintain polymorphisms for survival against infections [3,4]. These selected pressures also lead to a hitch-hiking effect that results in the accumulation of many additional SNPs around the Mhc gene [30]. Namely, the DNA segments affected by hitch-hiking have arisen due to the accumulated effect of overdominant selection and balancing selection. In the HLA region, HLA-A, -B, -DR/DQ and -DP are thought to have been affected by hitch-hiking and associated with several diseases, such as IDDM, rheumatoid arthritis and psoriasis vulgaris [30,40]. The hitch-hiking effect was also observed in Gado-B region [Shiina et al, Unpublished data]. Because the CojaIIBs have developed at a faster evolutionary speed than the BLBs (Fig. 2), the CojaIIB region is likely to be also effected by hitch-hiking. If a harmful variation is generated around the Mhc gene via the hitch-hiking effect, then it is likely to be selected against by genetic recombination.
A serious problem for disease mapping in the Gado-B region is that genetic recombination within the BF, BL and BG loci is rarely observed under experimental conditions, Therefore, Gado-B haplotypes encompassing alleles in all three loci are the units of inheritance most often considered in relating the Gado-B complex to immunity and disease responses [5]. On the other hand, the quail appears to be relatively more resistant than the chicken to many viral diseases [41]. In addition, the quail is thought to play an important role in the evolution of influenza viruses by acting as an intermediate host in which avian influenza viruses can be amplified and transmitted to other animal species [42]. Since the Coja region has the duplication and divergence of CojaIIBs (Figs. 2, 3, Additional file 1), the Coja region may be a superior system for the selection of beneficial variations enabling more antigen presentation ability than the Gado-B region. Therefore, to identify the disease genetic factors associated with the Gado-B region, comparative genomic and disease mapping analysis of the Coja region is also important.
Conclusion
In conclusion, we characterized the genetic and genomic features of the CojaIIB region and obtained the following three main findings. Firstly, one to six transcribed CojaIIB loci were identified in each Coja haplotype, of which one or two of them was the major coding locus. The major CojaIIB genes, except for CojaII-17 of HT3, were located within the Tapasin-BRD2 segment. In contrast to the quail, the chicken has one major and one minor MhcIIB. Secondly, phylogenetic and evolutionary analyses suggest that the major transcribed CojaIIBs were organized by independent trans-segmental duplications involving non-Mhc genes and/or transpositions between major and minor CojaIIBs. Consequently, the CojaIIBs have a relatively faster evolutionary speed than the BLBs. In contrast to the quail, the chicken BLB2 alleles have been generated from the ancestral BLB2 probably since the separation of chickens and quails from their common ancestor. Thirdly, four types of genome rearrangement elements (remodeling repeats) composed of tandem repeats were identified within the 4 ~ 11 kb haplotypic Tapasin-BRD2 segment, and the quail has far more repeat numbers for rearrangement events than the chicken that displays strong linkage disequilibrium [19-21]. Taken together, these three main findings support the view that the genomic diversity of the CojaIIB region has been generated by duplications and gene transpositions via the candidate rearrangement elements along with a fast evolutionary speed in adapting to environmental conditions and pathogens during the migratory history of the quail after its divergence from the chicken. It is evident from our study that the quail MhcIIB region has a much more flexible genomic structure than the chicken for generating greater Mhc diversity.
Methods
Quail
Blood samples were collected from 48 randomly selected quails maintained at the Tokyo University of Agriculture [23,43]. The blood collection and animal studies were conducted in accordance with the Guidelines for Animal Experiments at the Tokyo University of Agriculture.
Genotyping and haplotype analyses
Three polymorphic markers, PM1 to PM3, were identified in the Mhc classIIB genomic region of the quail (Figure 1) and were used as markers in this study for an initial haplotype analysis [14] (Tables 1, 2) either by PCR and sequence based typing or by PCR and RFLP analysis. PM1 is a mini-satellite marker composed of two different types of repetitive sequences and located in the Tapasin gene; PM2 is Coja-DBB1 locus genotyping marker; and PM3 is composed of two SNP markers located within the Coja-DMB2 gene (Table 1). Five different PM1 alleles were first identified by sequencing and then correlated with the length of their PCR products. The PCR product sizes of the PM1 alleles were then used to identify and define the alleles in the individual samples without the need for further sequencing. The PM1 alleles were designated as "6–10", "8–12", "9–12", "10–14" and "18–14" based on the length of the repeat units. For example, the "6–10" allele corresponds to the sequence TTCCTATGGGGGCTGTAGGGTGGATGGGACTGGGTGGTA6 – ATGAT10. Five PM2 alleles (Table 2) amplified by PCR were detected by a sequence based typing method of the PCR products and designated as Coja-DBB1*01 (alias "*01"), "*02", "*03", "*04" and "*05". The three PM3 alleles designated as alleles "G-C", "G-G" and "A-C" were detected by a PCR-RFLP method using DraIII and HinfI (Table 1) to identify the SNPs "G/A" and "G/C" at the two SNP sites that are apart by 194 bp. Haplotype prediction was performed by PHASE and Arlequin programs [44,45]. Maximum-likelihood haplotype frequencies were predicted by a Bayesian statistical method of the PHASE program and an EM algorithm of the Arlequin program.
RT-PCR, cloning cDNA and sequence determination
Total RNA was isolated from the peripheral blood of six quails identified by the numbers302, 311, 312, 321, 322 and 323 and having the haplotypes (HTs) 1/1, 2/3, 1/3, 1/4, 4/5, and 2/4 respectively. The TRIzol reagent was used to isolate the total RNA as described by the manufacturer (Invitrogen, Groningen, Netherlands). Total RNA (0.2 ug) was synthesized to cDNA by using First Strand cDNA Synthesis Kit (Rever Tra Ace-α-) and the oligo (dT) primer method (TOYOBO, Osaka, Japan). The CojaIIB specific primers were designed to amplify the exons 1 – 3 (amplified size: from 411 bp to 441 bp) and to detect polymorphisms on hyper variable β1 domain (exon 2) by RT-PCR amplification with the sense primer (C2BNO2: 5'-GAGTGCCACTACCTGAACGGCACCGAGG-3') and the anti-sense primer (C2BNO5: 5'-GCGCCAGGAAGACGAGCCCCAGCAC-3'). These primer sequences were perfectly matched with all CojaIIB genes on the Coja haplotype 1 to 5 in this study, providing confidence that the primers will amplify all the known CojaIIB genes with the same efficiency. cDNA (10 ng) was amplified by PCR with 2.5 units of GeneTaq NT polymerase (Nippon Gene, Toyama, Japan), using the thermal cycler GeneAmp PCR system 9700. The reaction mixture (10 ul) was subjected to 30 cycles of 30 sec at 96°C, 30 sec at 65°C, and 30 sec at 72°C. The RT-PCR products were cloned into the pGEM-T Easy vector with the TA cloning kit according to the protocol provided by the manufacturer (Promega, Madison, WI, USA) and sequenced using the ABI3100 genetic analyzer (Applied Biosystems, Foster City, CA, USA) in accordance with the protocol of BigDye terminator method. To avoid PCR and sequencing artifacts generated by polymerase errors, 96 clones per individual were sequenced.
Construction and screening of cosmid libraries
Genomic DNAs for constructing the cosmid libraries were isolated from the red blood cells of two Coja haplotype heterozygote quails (311 and 322), having HTs 2/3 and 4/5, by the Saponin-NaCl method [43]. The libraries were constructed by the SuperCos 1/Gigapack XL cloning kit according to the manufacturer's protocol (Stratagene, La Jolla, CA, USA). Approximately 5 × 105 independent colonies derived from these libraries were screened by using PCR products as hybridization probes obtained from the 527 ~ 586 bp CojaIIB fragment generated by the sense primer C2BNO2 and the anti-sense primer C2BNO5 already described above in the RT-PCR section; the 276 bp Tapasin fragment generated by sense primer Tapasin/e5-1 (5'-CCCAAAGAACCTGGTGGTGA-3') and anti-sense primer Tapasin/e5-4 (5'-AATGACCGTGGGTGACAA-3') and the 134 bp BRD2 fragment produced by the sense primer RING3/e4-1 (5'-GCAGCGGAGTGCAGAGACTT-3') and anti-sense primer RING3/e4-3 (5'-CGAGTGCCAGCTGTCTCCTC-3').
Genomic sequencing strategy
The DNA of CojaIIB, Tapasin and BRD2 positive cosmids was sequenced by the shotgun method [46]. Individual sequences were minimally edited to remove vector sequences, and assembled into a contig using the Sequencher software (Gene Codes Co., Ann Arbor, MI, USA). Remaining gaps or ambiguous nucleotides were determined by the direct sequencing of the PCR products obtained with appropriate PCR primers or by nucleotide sequence determination of additional shotgun clones.
Sequence analyses
Nucleotide similarities among the sequences were calculated by using the GENETYX-MAC software ver 11.0 (Software Development Co. Ltd., Tokyo, Japan). Dot-matrix analysis was performed by using HARRPLOT Ver. 2.0 as part of the GENETYX package. Multiple sequence alignments were created using the ClustalW Sequence Alignment program at DDBJ [47]. The phylogenetic tree was constructed by neighbor-joining method of the Molecular Evolution Genetics Analysis (MEGA3.1) [48].
Authors' contributions
KHos, TS, HI and KHan participated in the design of this study. KHos and TS constructed the quail cosmid libraries. KHos, TS, SSuz, MT, SShi, SI, HH, YY, HI and KHan participated in sequencing and contig assembly of the cosmid clones and data analysis. KHos, TS, HI, JKK and KHan prepared this manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Alignment of transcribed CojaIIBs for phylogenetic analysis. the nucleotide sequences of the β1 extracellular domain regions (exon 2) (270 nucleotides in length) of MhcIIB genes for the 16 CojaIIBs the chicken haplotypic BLB sequences on B2 B4 B12 B14 B15 B19 and B21.
Dot-matrix analysis among five Coja haplotypes (A ~ J) between quail and chicken (K) and between chicken B12 and B21 (L). Dot plot comparisons shows HT1 vs HT2 (A) HT1 vs HT3 (B) HT1 vs HT4 (C) HT1 vs HT5 (D) HT2 vs HT3 (E) HT2 vs HT4 (F) HT2 vs HT5 (G) HT3 vs HT4 (H) HT3 vs HT5(I) HT4 vs HT5 (J) HT2 vs chicken B12 (K) and chicken B12 vs B21 (L). Numbers and blue circles in the images show the location of genome candidate remodeling (rearrangement) factors with the numbers 1 to 3 representing TB1 – TB3 respectively as outlined in Table 6. Gray background shows the gap within the segments that was determined by sequencing.
Acknowledgments
Acknowledgements
The work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Animal Genome Research Project of the Ministry of Agriculture, Forestry and Fisheries of Japan and the Advanced Research Project Type A, Tokyo University of Agriculture, No.02, 2006.
Contributor Information
Kazuyoshi Hosomichi, Email: hoso@is.icc.u-tokai.ac.jp.
Takashi Shiina, Email: tshiina@is.icc.u-tokai.ac.jp.
Shingo Suzuki, Email: 53050010@nodai.ac.jp.
Masayuki Tanaka, Email: matanaka@jbirc.aist.go.jp.
Sayoko Shimizu, Email: sshimizu@is.icc.u-tokai.ac.jp.
Shigehisa Iwamoto, Email: 54050001@nodai.ac.jp.
Hiromi Hara, Email: hiromi-h@nodai.ac.jp.
Yutaka Yoshida, Email: yoshida@nodai.ac.jp.
Jerzy K Kulski, Email: jkulski@ccg.murdoch.edu.au.
Hidetoshi Inoko, Email: hinoko@is.icc.u-tokai.ac.jp.
Kei Hanzawa, Email: khanzawa@nodai.ac.jp.
References
- Klein J. Antigen-major histocompatibility complex-T cell receptors: inquiries into the immunological menage a trois. Immunol Res. 1986;5:173–190. doi: 10.1007/BF02919199. [DOI] [PubMed] [Google Scholar]
- Bourlet Y, Behar G, Guillemot F, Frechin N, Billault A, Chausse AM, Zoorob R, Auffray C. Isolation of chicken major histocompatibility complex class II (B-L) beta chain sequences: comparison with mammalian beta chains and expression in lymphoid organs. EMBO J. 1988;7:1031–1039. doi: 10.1002/j.1460-2075.1988.tb02910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Satta Y, Klein J. Polymorphism and balancing selection at major histocompatibility complex loci. Genetics. 1992;130:925–938. doi: 10.1093/genetics/130.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MM, Bacon LD, Hala K, Hunt HD, Ewald SJ, Kaufman J, Zoorob R, Briles WE. 2004 Nomenclature for the chicken major histocompatibility (B and Y) complex. Immunogenetics. 2004;56:261–279. doi: 10.1007/s00251-004-0682-1. [DOI] [PubMed] [Google Scholar]
- Briles WE, Goto RM, Auffray C, Miller MM. A polymorphic system related to but genetically independent of the chicken major histocompatibility complex. Immunogenetics. 1993;37:408–414. doi: 10.1007/BF00222464. [DOI] [PubMed] [Google Scholar]
- Miller MM, Goto R, Bernot A, Zoorob R, Auffray C, Bumstead N, Briles WE. Two Mhc class I and two Mhc class II genes map to the chicken Rfp-Y system outside the B complex. Proc Natl Acad Sci USA. 1994;91:4397–4401. doi: 10.1073/pnas.91.10.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillon V, Zoorob R, Yerle M, Auffray C, Vignal A. Mapping of the genetically independent chicken major histocompatibility complexes B and RFP-Y to the same microchromosome by two-color fluorescent in situ hybridization. Cytogenet Cell Genet. 1996;75:7–9. doi: 10.1159/000134445. [DOI] [PubMed] [Google Scholar]
- Miller MM, Goto RM, Taylor RL, Jr, Zoorob R, Auffray C, Briles RW, Briles WE, Bloom SE. Assignment of Rfp-Y to the chicken major histocompatibility complex/NOR microchromosome and evidence for high-frequency recombination associated with the nucleolar organizer region. Proc Natl Acad Sci USA. 1996;93:3958–3962. doi: 10.1073/pnas.93.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J, Milne S, Gobel TW, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S. The chicken B locus is a minimal essential major histocompatibility complex. Nature. 1999;401:923–925. doi: 10.1038/44856. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Ruby T, Bed'Hom B, Wittzell H, Morin V, Oudin A, Zoorob R. Characterisation of a cluster of TRIM-B30.2 genes in the chicken MHC B locus. Immunogenetics. 2005;57:116–128. doi: 10.1007/s00251-005-0770-x. [DOI] [PubMed] [Google Scholar]
- Shiina T, Hosomichi K, Hanzawa K. Comparative genomics of the poultry major histocompatibility complex. Anim Sci J. 2004;77:151–162. doi: 10.1111/j.1740-0929.2006.00333.x. [DOI] [Google Scholar]
- Shiina T, Shimizu S, Hosomichi K, Kohara S, Watanabe S, Hanzawa K, Beck S, Kulski JK, Inoko H. Comparative Genomic Analysis of Two Avian (Quail and Chicken) MHC Regions. J Immunol. 2004;172:6751–6763. doi: 10.4049/jimmunol.172.11.6751. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H. Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev. 2002;190:95–122. doi: 10.1034/j.1600-065X.2002.19008.x. [DOI] [PubMed] [Google Scholar]
- Collins WM, Briles WE, Zsigray RM, Dunlop WR, Corbett AC, Clark KK, Marks JL, McGrail1 TP. The B locus (MHC) in the chicken: Association with the fate of RSV-induced tumors. Immunogenetics. 1977;5:333–343. doi: 10.1007/BF01570490. [DOI] [Google Scholar]
- Briles WE, Stone HA, Cole RK. Marek's disease: effects of B histocompatibility alloalleles in resistant and susceptible chicken lines. Science. 1977;195:193–195. doi: 10.1126/science.831269. [DOI] [PubMed] [Google Scholar]
- Longenecker BM, Gallatin WM. Genetic control of resistance to Marek's disease. IARC Sci Publ. 1978:845–850. [PubMed] [Google Scholar]
- Simonsen M, Crone M, Koch C, Hala K. The MHC haplotypes of the chicken. Immunogenetics. 1982;16:513–532. doi: 10.1007/BF00372021. [DOI] [PubMed] [Google Scholar]
- Hala K, Chausse AM, Bourlet Y, Lassila O, Hasler V, Auffray C. Attempt to detect recombination between B-F and B-L genes within the chicken B complex by serological typing, in vitro MLR, and RFLP analyses. Immunogenetics. 1988;28:433–438. doi: 10.1007/BF00355375. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Volk H, Wallny HJ. A "minimal essential Mhc" and an "unrecognized Mhc": two extremes in selection for polymorphism. Immunol Rev. 1995;143:63–88. doi: 10.1111/j.1600-065X.1995.tb00670.x. [DOI] [PubMed] [Google Scholar]
- Wilcox FH, Clark CE. Chicken-quail hybrids. J Hered. 1961;52:167–170. [Google Scholar]
- Watanabe S, Nagayama F. Studies on the serum IgG level in Japanese quail. Japanese Poultry Science. 1979;16:59–64. [Google Scholar]
- Inooka S, Takahashi S, Takahashi H, Mizuma Y. Immunological traits in generations 7 to 12 of two lines of Japanese quail selected for high or low antibody response to Newcastle disease virus. Poult Sci. 1984;63:1298–1302. doi: 10.3382/ps.0631298. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Shiina T, Hosomichi K, Takahashi S, Koyama T, Onodera T, Kulski JK, Inoko H. MHC class IIB gene sequences and expression in quails (Coturnix japonica) selected for high and low antibody responses. Immunogenetics. 2004;56:280–291. doi: 10.1007/s00251-004-0690-1. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Inooka S, Mizuma Y. Selective breeding for high and low antibody responses to inactivated Newcastle disease virus in Japanese quails. Poult Sci. 1984;63:595–599. doi: 10.3382/ps.0630595. [DOI] [PubMed] [Google Scholar]
- Pradhan HK, Mohanty GC, Mukit A. Marek's disease in Japanese quails (Coturnix coturnix japonica): a study of natural cases. Avian Dis. 1985;29:575–582. doi: 10.2307/1590648. [DOI] [PubMed] [Google Scholar]
- Shiina T, Shimizu C, Oka A, Teraoka Y, Imanishi T, Gojobori T, Hanzawa K, Watanabe S, Inoko H. Gene organization of the quail major histocompatibility complex (MhcCoja) class I gene region. Immunogenetics. 1999;49:384–394. doi: 10.1007/s002510050511. [DOI] [PubMed] [Google Scholar]
- Shiina T, Oka A, Imanishi T, Hanzawa K, Gojobori T, Watanabe S, Inoko H. Multiple class I loci are expressed in the quail MHC. Immunogenetics. 1999;49:456–460. doi: 10.1007/s002510050519. [DOI] [PubMed] [Google Scholar]
- Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, Takasu M, Anzai T, Kulski JK, Kikkawa E, Naruse T, Kimura N, Yanagiya K, Watanabe A, Hosomichi K, Kohara S, Iwamoto C, Umehara Y, Meyer A, Wanner V, Sano K, Macquin C, Ikeo K, Tokunaga K, Gojobori T, Inoko H, Bahram S. Rapid Evolution of MHC Class I Genes in Primates Generates New Disease Alleles in Man Via Hitchhiking Diversity. Genetics. 2006;173:1555–1570. doi: 10.1534/genetics.106.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corell A, Morales P, Varela P, Paz-Artal E, Martin-Villa JM, Martinez-Laso J, Arnaiz-Villena A. Allelic diversity at the primate major histocompatibility complex DRB6 locus. Immunogenetics. 1992;36:33–38. doi: 10.1007/BF00209290. [DOI] [PubMed] [Google Scholar]
- Matsuo MY, Asakawa S, Shimizu N, Kimura H, Nonaka M. Nucleotide sequence of the MHC class I genomic region of a teleost the medaka (Oryzias latipes) Immunogenetics. 2002;53:930–940. doi: 10.1007/s00251-001-0427-3. [DOI] [PubMed] [Google Scholar]
- Shiina T, Dijkstra JM, Shimizu S, Watanabe A, Yanagiya K, Kiryu I, Fujiwara A, Nishida-Umehara C, Kaba Y, Hirono I, Yoshiura Y, Aoki T, Inoko H, Kulski JK, Ototake M. Interchromosomal duplication of major histocompatibility complex class I regions in rainbow trout (Oncorhynchus mykiss) a species with a presumably recent tetraploid ancestry. Immunogenetics. 2005;56:878–893. doi: 10.1007/s00251-004-0755-1. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Goetz W, Hossain MZ, Nonaka M, Flajnik MF. Ancestral organization of the MHC revealed in the amphibian Xenopus. J Immunol. 2006;176:3674–3685. doi: 10.4049/jimmunol.176.6.3674. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Gaudieri S, Bellgard M, Balmer L, Giles K, Inoko H, Dawkins RL. The evolution of MHC diversity by segmental duplication and transposition of retroelements. J Mol Evol. 1997;45:599–609. doi: 10.1007/PL00006264. [DOI] [PubMed] [Google Scholar]
- Shiina T, Tamiya G, Oka A, Takishima N, Yamagata T, Kikkawa E, Iwata K, Tomizawa M, Okuaki N, Kuwano Y, Watanabe K, Fukuzumi Y, Itakura S, Sugawara C, Ono A, Yamazaki M, Tashiro H, Ando A, Ikemura T, Soeda E, Kimura M, Bahram S, Inoko H. Molecular dynamics of MHC genesis unraveled by sequence analysis of the 1796938-bp HLA class I region. Proc Natl Acad Sci USA. 1999;96:13282–13287. doi: 10.1073/pnas.96.23.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D. Object-oriented transcription factors database (ooTFD) Nucleic Acids Res. 2000;28:308–310. doi: 10.1093/nar/28.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroishi T, Hanzawa N, Sagai T, Ishiura M, Gojobori T, Steinmetz M, Moriwaki K. Recombinational hotspot specific to female meiosis in the mouse major histocompatibility complex. Immunogenetics. 1990;31:79–88. doi: 10.1007/BF00661217. [DOI] [PubMed] [Google Scholar]
- Crawford DC, Bhangale T, Li N, Hellenthal G, Rieder MJ, Nickerson DA, Stephens M. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DR, Lim W, Seiler JP, Yi G, Peiris M, Shortridge KF, Webster RG. Role of quail in the interspecies transmission of H9 influenza A viruses: molecular changes on HA that correspond to adaptation from ducks to chickens. J Virol. 2003;77:3148–3156. doi: 10.1128/JVI.77.5.3148-3156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnamohan N. The management of Japanese quail and their use in virological research: a review. Vet Res Commun. 1985;9:1–14. doi: 10.1007/BF02215123. [DOI] [PubMed] [Google Scholar]
- Shiina T, Ando A, Imanishi T, Kawata H, Hanzawa K, Gojobori T, Inoko H, Watanabe S. Isolation and characterization of cDNA clones for Japanese quail (Coturnix japonica) major histocompatibility complex (MhcCoja) class I molecules. Immunogenetics. 1995;42:213–216. doi: 10.1007/BF00191227. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Tamiya G, Oka A, Yamagata T, Yamagata N, Kikkawa E, Goto K, Mizuki N, Watanabe K, Fukuzumi Y, Taguchi S, Sugawara C, Ono A, Chen L, Yamazaki M, Tashiro H, Ando A, Ikemura T, Kimura M, Inoko H. Nucleotide sequencing analysis of the 146-kilobase segment around the IkBL and MICA genes at the centromeric end of the HLA class I region. Genomics. 1998;47:372–382. doi: 10.1006/geno.1997.5114. [DOI] [PubMed] [Google Scholar]
- DDBJ http://www.ddbj.nig.ac.jp/search/Welcome-e.html
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of transcribed CojaIIBs for phylogenetic analysis. the nucleotide sequences of the β1 extracellular domain regions (exon 2) (270 nucleotides in length) of MhcIIB genes for the 16 CojaIIBs the chicken haplotypic BLB sequences on B2 B4 B12 B14 B15 B19 and B21.
Dot-matrix analysis among five Coja haplotypes (A ~ J) between quail and chicken (K) and between chicken B12 and B21 (L). Dot plot comparisons shows HT1 vs HT2 (A) HT1 vs HT3 (B) HT1 vs HT4 (C) HT1 vs HT5 (D) HT2 vs HT3 (E) HT2 vs HT4 (F) HT2 vs HT5 (G) HT3 vs HT4 (H) HT3 vs HT5(I) HT4 vs HT5 (J) HT2 vs chicken B12 (K) and chicken B12 vs B21 (L). Numbers and blue circles in the images show the location of genome candidate remodeling (rearrangement) factors with the numbers 1 to 3 representing TB1 – TB3 respectively as outlined in Table 6. Gray background shows the gap within the segments that was determined by sequencing.


