Abstract
Aim: Several studies have investigated the expression of the cytokeratins (CKs), vimentin, the epithelial growth factor receptor (EGFR), the oestrogen receptor (ER), and the progesterone receptor (PgR), in breast cancer, but no study has directly compared p53 mutations with these phenotypic and differentiation markers in the same case. The present study was designed to provide some of this information.
Methods: The expression of the p53 and bcl-2 proteins was evaluated by immunohistochemistry in relation to phenotypic characteristics and cellular kinetic parameters (mitotic index and apoptotic index) in 37 cases of ductal carcinoma in situ (DCIS) and 27 cases of infiltrating ductal carcinoma (IDC) of the breast. In addition, p53 gene mutation was examined by polymerase chain reaction single strand conformation polymorphism analysis (SSCP).
Results: Thirteen cases (eight DCIS and five IDC) showed expression of CK8, CK14, CK18, vimentin, and EGFR, consistent with a stem cell phenotype, whereas 44 cases (27 DCIS and 17 IDC) showed expression of CK8 and CK1, weak or negative expression of CK18, but were negative for vimentin and EGFR, consistent with a luminal cell phenotype. DCIS and IDC cases with a stem cell phenotype were ER/PgR negative and intermediately or poorly differentiated. In contrast, the cases with luminal cell phenotype were ER/PgR positive and well or intermediately differentiated. In addition, intermediately or poorly differentiated cases with a stem cell phenotype showed higher proliferative activity (per cent of MIB-l positive cells) than did intermediately or well differentiated cases with a luminal cell phenotype. Both DCIS and IDC cases with a stem cell phenotype were p53 positive and bcl-2 negative by immunohistochemistry. In IDC, p53 expression was associated with a reduction of both mitotic index and apoptotic index compared with DCIS. Most of the tumours showing a more differentiated phenotype (luminal) were p53 negative and bcl-2 positive. In these cases, cell kinetic parameters increased from DCIS to IDC. These data suggest the existence of subsets of DCIS and IDC that, because of their phenotypic characteristics, could be derived from subpopulations of normal breast cells having different control mechanisms of cell proliferation and neoplastic progression.
Conclusions: These results are compatible with the hypothesis that the phenotype of the cell of origin constrains both tumour phenotype and the choice of genetic events; however, the occurrence of p53 mutants by chance during neoplastic transformation cannot be excluded.
Keywords: ductal carcinoma in situ, infiltrating ductal carcinoma, cell kinetics, p53, bcl-2
Abnormalities of the p53 tumour suppressor gene are among the most frequent molecular events in human and animal neoplasia.1 It is now accepted that the inactivation of this gene, as a result of mutation, is a key step in neoplastic transformation and progression.2,3 This effect is usually related to the loss of p53 activated cell death, growth arrest, and/or control of genomic stability.2 However, it is not yet completely clear which of the many properties of p53 are particularly important in oncogenesis.4,5 In addition, the p53 activation system may be influenced by cell type and specific tissue effects.2 About 30–50% of breast cancers carry a mutant p53 gene and show p53 protein expression by immunohistochemistry.6
The expression of this oncoprotein in breast cancers seems to be related to poor prognosis associated with a high histological grade, epidermal growth factor receptor (EGFR) positivity, and bcl-2 and oestrogen receptor (ER) negativity.7–16 However, it should be noted that clinical pathological studies also report the coexpression of cytokeratin (CK) and vimentin in breast cancer as a more aggressive phenotype associated with poor prognosis.17,18 This particular phenotype has been detected in a subset of normal breast epithelial cells that grows out of mammoplasty cultures, after most of the early passage cells have senesced.19,20 The high proliferative capacity of these cells, together with this uncommitted differentiation state, has led many observers to regard them as being derived from a small “stem cell” pool present in the basal layer of the normal gland.21 In contrast, most normal breast epithelial cells present in the intact gland and in early passage mammoplasty cultures express CK8, CK18 (weakly), and CK19, but not vimentin.20
“It is now accepted that the inactivation of the p53 gene, as a result of mutation, is a key step in neoplastic transformation and progression”
Although several studies have investigated the expression of CKs and vimentin, EGFR, ER, and the progesterone receptor (PgR) in breast cancer,17,18,22–30 to our knowledge, no study has directly compared p53 mutations with these phenotypic and differentiation markers in the same case. Our present study was designed to provide some of this information. We therefore analysed the expression of p53 (wild-type and mutated form) and bcl-2 proteins in relation to phenotypic characteristics in 37 cases of ductal carcinoma in situ (DCIS) and 27 cases of infiltrating ductal carcinoma (IDC) of the breast. Because these oncoproteins are involved in the regulation of cell proliferation and cell death, we also assessed, in each case, the proliferative activity, and mitotic and apoptotic indices (MI and AI, respectively).
MATERIALS AND METHODS
Data collection
Formalin fixed, paraffin wax embedded tissue samples of 64 serial archival cases of breast carcinoma (37 cases of DCIS and 27 cases of IDC) were used. None of the DCIS cases showed any adjacent areas of invasive carcinoma. Nineteen IDC cases showed adjacent areas of in situ carcinoma. All patients were diagnosed at the Institute of Pathological Anatomy and Histology, University of Siena. The patients' ages ranged from 29 to 85 years, with a median age of 65. All patients had been treated with partial or modified radical mastectomy and axillary node dissection, with no previous chemotherapy or radiotherapy. Haematoxylin and eosin stained sections of all cases were reviewed and classified according to the common histological criteria of the World Health Organisation classification.31 The pathological stage was assessed according to the criteria established by the International Union Against Cancer.32
Immunohistochemistry
Paraffin wax sections were cut to a thicknesses of 5 μm, immersed in citric acid buffer (pH 6.0) and incubated twice in a microwave oven at 750 W power for five minutes each. Table 1 details the monoclonal antibodies used. The sections were immunostained by the APAAP complex method (Dako, Glostrup, Denmark). Antibody expression was evaluated in each case by two experienced pathologists (TM, FF), independently, and consensus was reached in every case. Cases were considered positive for CK8, CK14, CK18, and vimentin when cytoplasmic staining was seen in all neoplastic cells. EGFR was only considered positive when membrane staining was seen. A semiquantitative score (±) was given for weak or focal positivity for CK18, vimentin, or EGFR.
Table 1.
Antibodies used for immunohistochemical reactions on paraffin wax sections of formalin fixed tissue
| Antigen/ antibody | Clone | Source |
| CK8 | 5D3 | Menarini Diagnostics (Florence, Italy) |
| CK18 | 5D3 | Menarini Diagnostics |
| CK14 | LL002 | Menarini Diagnostics |
| Vimentin | V9 | Dako (Milan, Italy) |
| EGFR | 31G7 | DBA |
| ER | ER1D5 | Immunotech (Marseille, France) |
| PgR | PR10A9 | Immunotech |
| MIB1 | MIB-1 | Immunotech |
| p53 | DO-7 | Immunotech |
| Bcl-2 | 124 | Dako (Milan, Italy) |
CK, cytokeratin; EGFR, epidermal growth factor receptor; ER, oestrogen receptor; PgR, progesterone receptor.
We considered tumours to be positive for p53 or bcl-2 expression when 10% or more of the cells counted were stained, but we made no attempt to quantify the intensity of immunostaining for each cell.33 Furthermore, we graded MIB-1, ER, and PgR expression as low or high when the percentages of positive cells were < 40% or ≥ 40%, respectively, based on the frequency histogram analysis. For negative controls, we replaced primary antibody with normal mouse serum.
In situ end labelling (ISEL)
For comparison with the morphologically registered apoptotic cells/bodies, the in situ end labelling (ISEL) technique, which visualises DNA fragmentation, was applied with minor modifications as reported previously.34 Negative controls were obtained by omitting the DNA polymerase in the labelling mix.
Cell counts
For each case, we first assessed the cellularity by determining the number of intact epithelial cells in each high power field (HPF; median, 1200; range, 642–1872) in 30 randomly chosen HPFs (HPF, 56 000 μm2/field), using an oil immersion objective (×l00). We then assessed the MI and AI by counting the percentages of neoplastic cells in mitosis and the number of apoptotic cells/bodies for each 100 neoplastic cells in an entire section for each case (∼ 100 HPFs). With this approach, the relative number of apoptotic cells/bodies corresponded well with the percentage of cells showing ISEL of the DNA strand breaks.34,35 The intra-observer and interobserver reproducibility of the counts was 95%.
Positively stained cells for all the antibodies used were counted in 30 randomly chosen HPFs and expressed as percentages of all epithelial cells. The slides were scored by two independent pathologists (CB, AVL), each performing repeated counts on the same section, but viewing different HPFs. To verify the reproducibility, the absence of any systematic differences between independent scorings was preliminarily tested with the Wilcoxon signed rank test for paired samples. No significant differences were found (p > 0.l). A good agreement between observers (little variance of the differences) was then shown by Pearson correlation analysis (r = 0.95; p < 10−14).
Molecular analysis: screening for p53 gene mutation (PCR-SSCP)
The method used in our study for identifying mutations in the p53 gene involved DNA amplification using the polymerase chain reaction (PCR), followed by identification of the mutated products using the single strand conformation polymorphism (SSCP) technique.
DNA was extracted from paraffin wax embedded tumour tissues following the procedure described previously.35 PCR of exons 5–10 was carried out in a 50 μl volume, consisting of 5× reaction buffer (300 mmol/litre Tris/HCl, 75 mmol/litre (NH4)2S04, 7.5 mmol/litre MgCl2 (pH 8.5)), dNTP mixture (final concentration 0.25 mmol/litre), 1.0 U cloned Thermus aquaticus DNA polymerase (Life Technologies, Milan, Italy), and p53 primers21,22 (final concentration 0.4 μmol/litre). PCR amplifications were run in a programmable thermal cycler (Eppendorf, Netheler, Germany) for one minute at 94°C, one minute at 56°C (exons 5 and 8) or 59°C (exons 6, 7, 9, and 10), and one minute at 72°C, followed by a final 10 minutes at 72°C. Non-isotopic SSCP analysis of PCR products was carried out using a GenPhor electrophoresis unit (Pharmacia Biotech, Uppsala, Sweden) on 12.5 % polyacrylamide gels (GeneGel Excel, 12.5/24 Kit; Pharmacia Biotech) at 400 V, 10°C, for 1.5 hours. Single stranded DNA fragments were stained with silver in an automated gel stainer (Hoefer Pharmacia Biotech).
Statistical analysis of scoring data
Sample mean and SD values were computed as descriptive statistics for the analysed variables (MI, AI, and MIB-1). Linear correlation analysis between these variables was performed by means of Pearson coefficient of correlation (r). A regression line of MI against AI was also computed. The Mann-Whitney test for independent samples was used to compare the differences between sample groups.
RESULTS
Expression of CK, vimentin, and EGFR in DCIS and IDC of the breast
Table 2 details the expression of CK8, CK18, CK14, vimentin, and EGFR in DCIS and IDC of the breast.
Table 2.
Expression of CKs, vimentin, and EGFR in DCIS and IDC
| CK8 | CK14 | CK18 | Vimentin | EGFR | Total case | |
| Stem cell phenotype | ||||||
| DCIS | + | + | + | + | + | 8 |
| IDC | + | + | + | + | + | 5 |
| Luminal cell phenotype | ||||||
| DCIS | + | + | +/− | − | − | 27 |
| IDC | + | + | +/− | − | − | 17 |
| Not classifiable | ||||||
| DCIS | + | + | + | +/− | +/− | 2 |
| IDC | + | + | + | +/− | +/− | 5 |
CK, cytokeratin; DCIS, ductal carcinoma in situ; EGFR, epidermal growth factor receptor; IDC, infiltrating ductal carcinoma.
Thirteen cases (eight DCIS and five IDC) showed expression of CK8, CK14, CK18, vimentin, and EGFR (fig 1). In contrast, 44 cases (27 DCIS and 17 IDC) showed expression of CK8, CK14, weak expression or negativity for CK18, and negativity for vimentin and EGFR (fig 2). The cases of IDC with adjacent in situ areas showed the same phenotype in both components. According to these phenotypic characteristics, our cases were considered to be of stem or luminal cell origin, respectively.
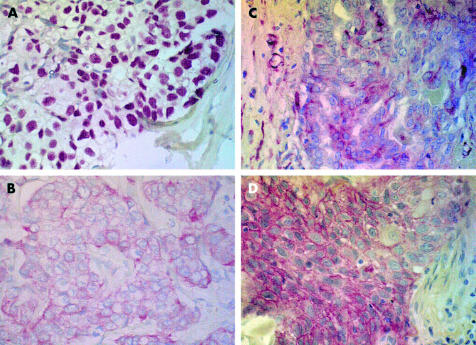
Figure 1.
Expression of (A) p53 protein in relation to the stem cell phenotype: (B) CK18+, (C) vimentin positive, and (D) epidermal growth factor positive.
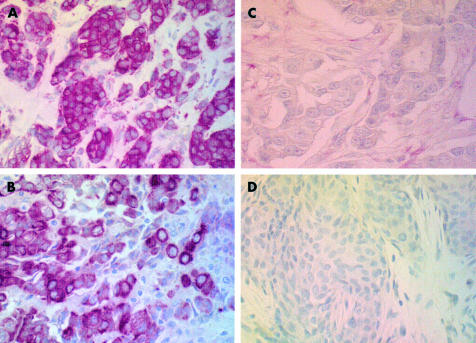
Figure 2.
Expression of (A) bcl-2 protein in relation to the luminal cell phenotype: (B) CK8+, (C) vimentin negative, and (D) epidermal growth factor negative.
Two DCIS and five IDC cases showed a phenotype pattern unrelated to either a luminal or stem cell origin.
Histopathological characteristics of DCIS and IDC of the breast and correlation with phenotype characteristics
The cases with a stem cell phenotype (CK8+, CK14+, CK18+, vimentin positive, EGFR+) were characterised by ER/PgR negativity; four cases (three DCIS and one IDC) were intermediately differentiated and nine cases (five DCIS and four IDC) were poorly differentiated. In contrast, the cases with a luminal cell phenotype (CK8+, CK14+, CK18+/−, vimentin negative, EGFR−) showed ER and PgR positivity; seven cases (four DCIS and two IDC) were well differentiated, 31 cases were intermediately differentiated (18 DCIS and 12 IDC), and one IDC was poorly differentiated. Furthermore, DCIS and IDC cases with a stem cell phenotype had much higher values of MIB-1 positive cells (mean, 63.5% and 69.95%, respectively) than cases with a luminal cell phenotype (mean, 14.96% and 18.9%, respectively; p < 0.00l).
Expression of p53 and bcl-2 in DCIS and IDC of the breast and correlation with phenotype characteristics
p53+ and bcl-2+ cells showed nuclear and cytoplasmic positivity, respectively, in a variable number of neoplastic cells in both DCIS and IDC. A significant inverse correlation between bcl-2 and p53 expression was detected—15 cases (eight cases of DCIS and seven cases of IDC) were p53+/bcl-2−, and 44 cases (27 cases of DCIS and 17 cases of IDC) were, on the contrary, p53−/bcl-2+. Only five cases were bcl2+ and p53+. No p53−/bcl-2− cases were observed.
Table 3 summarises the expression of p53 and bcl-2 in DCIS and IDC in relation to phenotypic characteristics. Thirteen cases (eight DCIS and five IDC cases) were p53+/bcl-2− and showed a phenotype consistent with a stem cell origin (CK8+, CK14+, CKl8+, vimentin positive, EGFR+). Only two p53+/bcl-2− cases showed a luminal cell phenotype. Forty four cases were p53−/bcl-2+ (27 DCIS and 17 IDC cases) and 37 showed a phenotype consistent with a luminal cell origin (CK8+, CK14+, CK18+/−, vimentin negative, EGFR−). The five p53+/bcl-2+ cases also showed a phenotype consistent with a luminal cell origin.
Table 3.
Expression of p53 and bcl-2 in DCIS and IDC
| Stem cell phenotype | Luminal cell phenotype | Not classifiable | ||||
| DCIS | IDC | DCIS | IDC | DCIS | IDC | |
| p53+/bcl-2− | 8 | 5 | 0 | 2 | 0 | 0 |
| p53−/bcl-2+ | 0 | 0 | 25 | 12 | 2 | 5 |
| p53+/bcl-2+ | 0 | 0 | 2 | 3 | 0 | 0 |
DCIS, ductal carcinoma in situ; IDC, infiltrating ductal carcinoma.
Mutational screening of p53
Thirty seven DCIS and 27 IDC samples were examined for mutation in exons 5 to 10 of the p53 gene by PCR-SSCP. By this method, no band shifts were observed in DCIS, either in p53− cases or in p53+ cases evaluated by immunohistochemistry; conversely, band shifts suggestive of point mutation were observed only in the 10 cases of IDC that were also positive for p53 protein by immunohistochemistry. No band shifts were detected in IDCs that were negative for p53 protein by immunohistochemistry.
Cellular kinetics of DCIS and IDC of the breast and correlation with phenotype characteristics
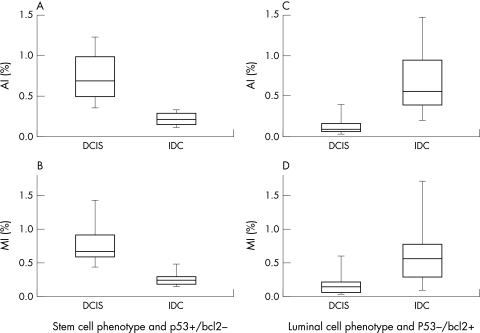
Figure 3 illustrates the cellular kinetic characteristics (MI and AI) of DCIS and IDC of the breast. In p53+/bcl-2− cases with a stem cell phenotype, there was a reduction of both MI and AI in IDC when compared with DCIS (p < 0.00l). The converse was seen for the p53−/bcl-2+ cases with a luminal cell phenotype (p < 0.001).
Figure 3.
Cellular kinetic characteristics (apoptotic index (AI) and mitotic index (MI)) of ductal carcinoma in situ (DCIS) and infiltrating ductal carcinoma (IDC): in p53+/bcl-2− cases with a stem cell phenotype there is a reduction of both AI (A) and MI (B) in IDC compared with DCIS (p < 0.001). In p53−/bcl-2+ cases with a luminal cell phenotype, AI (C) and MI (D) increased from DCIS to IDC (p < 0.001).
DISCUSSION
In our study we analysed the expression of p53 (wild-type and mutated form) and bcl-2 proteins in relation to the phenotypic and cellular kinetic characteristics of 37 DCIS and 27 IDC samples from the breast. The phenotypic characteristics of our cases suggest a different cell origin among breast cancers. In fact, 13 cases (eight DCIS and five IDC cases) showed expression of CK8, CK14, CK18, vimentin, and EGFR, consistent with a stem cell origin; 44 cases (27 DCIS and 17 IDC) showed expression of CK8 and CK14, weak expression or negativity for CK18, and negativity for vimentin and EGFR, consistent with a luminal cell origin. Two DCIS and five IDC cases showed a phenotypic pattern unrelated to either a luminal cell or stem cell origin. The interpretation of these results is of course speculative, because these cases may have lost their phenotype or may be derived from yet another precursor cell.
“Our results nevertheless suggest that p53 was wild-type in ductal in situ carcinoma cases and mutated in infiltrating ductal carcinoma cases because the SSCP technique has a sensitivity and specificity of approximately 90%
Both DCIS and IDC cases with a stem cell phenotype were p53+/bcl-2− by immunohistochemistry. Among these cases, the screening for p53 mutations with SSCP analysis showed a band shift suggesting a point mutation in IDC, whereas no band shift was detected in DCIS. Without sequencing the PCR products we cannot completely exclude either false negative cases, because we have examined only a limited number of p53 exons, or false positive cases for the presence of polymorphisms.36 With this restriction in mind, our results nevertheless suggest that p53 was wild-type in DCIS cases and mutated in IDC cases because the SSCP technique has a sensitivity and specificity of approximately 90%.37 Interestingly, when compared with DCIS, p53 expression in IDC was associated with a reduction of both MI and AI. Abundant apoptosis in DCIS has been identified independent of p53 immunostaining.38 In this last group, the expression of p53 might well reflect the presence of a wild-type protein, as a physiological response to DNA damage.2–4 Conversely, the p53 protein in IDC has probably lost its physiological function to induce cell death. It is well known that reduced apoptosis may lead to a shift in tissue kinetics towards increased cell numbers, and also to the preservation of genetically aberrant cells, thereby favouring neoplastic progression.39,40 In particular, this model has been observed in poorly differentiated carcinomas of the breast.41 In our series, the IDC cases with p53 mutation were poorly or moderately differentiated tumours, ER and PgR negative, and showed significantly higher numbers of MIB-1 positive cells. The high proliferative activity of these tumours, as compared with the reduction of MI, may also be linked to inactivation of p53, which leads to the abrogation of the G1/S block and/or the inhibition of G2 exit.42 The presence of “proliferation associated” proteins is not necessarily an indication that the respective cells will successfully terminate the cell cycle and divide. An alteration of cell cycle progression may be present in such cases and the neoplastic growth may be the result of a reduction of cell death rather than increased cell division. The identification of such a kinetic pattern may be of some prognostic value because high metastatic potential has been shown to be associated with increased resistance to apoptosis.43
Most of the tumours showing a more differentiated phenotype (luminal) were p53−/bcl-2+. In these cases, cell kinetic parameters increased from DCIS to IDC. Similar kinetic characteristics have been observed in well differentiated breast tumours.41 The increased values of AI and MI from DCIS to IDC in such cases could be related to an alteration of cell growth control, probably as a result of other genetic lesions involving the retinoblastoma pathway.44 The coexistence of high percentages of bcl-2+ cells and increased AI could tentatively be explained by bcl-2 independent pathways to programmed cell death.45
Take home messages.
Thirteen cases (eight ductal carcinoma in situ (DCIS) and five infiltrating ductal carcinoma (IDC)) showed expression characteristics consistent with a stem cell phenotype, whereas 44 cases (27 DCIS and 17 IDC) showed characteristics consistent with a luminal cell phenotype
Most of the tumours showing a more differentiated phenotype (luminal) were p53−/bcl-2+. In these cases, cell kinetic parameters increased from DCIS to IDC
These data suggest the existence of subsets of DCIS and IDC that, because of their phenotypic characteristics, could be derived from subpopulations of normal breast cells having different control mechanisms of cell proliferation and neoplastic progression
However, the occurrence of p53 mutants by chance during neoplastic transformation cannot be excluded and further studies are warranted
In conclusion, our data suggests the existence of subsets of DCIS and IDC that, because of their phenotypic characteristics, could be derived from subpopulations of normal breast cells, which probably use different control mechanisms of cell proliferation and neoplastic progression—p53 dependent in the cases with a stem cell phenotype and p53 independent in the cases with a luminal cell phenotype. In fact, the liability of a given tumour to exhibit p53 mutation seems to be predetermined by the nature of its cell of origin, depending more specifically to what extent wild-type p53 forms a rate limiting step in the control of the proliferating life span in that cell.46,47
“The coexistence of high percentages of bcl-2+ cells and increased apoptotic index could tentatively be explained by bcl-2 independent pathways to programmed cell death”
Our results are compatible with the hypothesis that the cell of origin constrains both tumour phenotype and choice of genetic events.47 However, the occurrence of p53 mutants by chance during neoplastic transformation cannot be excluded, as is suggested by the finding of p53+/bcl-2+ cases. However, it must be remembered that in situ and invasive breast lesions were taken from different patients, and that none of the patients with an in situ lesion had adjacent invasive carcinoma. Therefore, our results should be interpreted with caution. More direct evidence of our hypothesis could be obtained from the study of patients with invasive breast cancer who had a previous breast biopsy that showed carcinoma in situ. We are presently gathering these cases and performing kinetic and molecular studies by capture laser microdissection.
Abbreviations
AI, apoptotic index
DCIS, ductal carcinoma in situ
EGFR, epidermal growth factor receptor
ER, oestrogen receptor
HPF, high power field
IDC, infiltrating ductal carcinoma
ISEL, in situ end labelling
MI, mitotic index
PCR, polymerase chain reaction
PgR, progesterone receptor
SSCP, single strand conformation polymorphism
REFERENCES
- 1.Hainaut P, Hernandez T, Robinson A, et al. IARC database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res 1998;26:205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prives C, Hall PA. The p53 pathway. J Pathol 1999;187:112–26. [DOI] [PubMed] [Google Scholar]
- 3.Greeblatt MS, Bennett WP, Hollstein M, et al. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 1994;54:4855–78. [PubMed] [Google Scholar]
- 4.Lane DP. The regulation of p53 function: Steiner award lecture. Int J Cancer 1994;57:623–7. [DOI] [PubMed] [Google Scholar]
- 5.Livingstone LR, White A, Sprouse J, et al. Altered cell cycle arrest and gene amplification potential accompany loss of wild type p53. Cell 1994;70:923–35. [DOI] [PubMed] [Google Scholar]
- 6.Sjogren S, Inganas M, Norbert T, et al. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst 1996;88:173–82. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann A, Blaszyk H, Kovach JS, et al. The molecular epidemiology of p53 gene mutations in human breast cancer. Trends Genet 1997;13:27–33. [DOI] [PubMed] [Google Scholar]
- 8.Barnes DM, Dublin EA, Fisher CJ, et al. Immunohistochemical detection of p53 protein in mammary carcinoma: an important new independent indicator of prognosis? Hum Pathol 1992;24:469–76. [DOI] [PubMed] [Google Scholar]
- 9.Isola J, Visakorpi T, Holli K, et al. Association of overexpression of tumor suppressor protein p53 with rapid cell proliferation and poor prognosis in node-negative breast cancer patients. J Natl Cancer Inst 1992;84:1109–14. [DOI] [PubMed] [Google Scholar]
- 10.Caleffi M, Teague MW, Jensen RA, et al. P53 gene mutations and steroid receptor status in breast cancer. Cancer 1994;73:2147–56. [DOI] [PubMed] [Google Scholar]
- 11.Bergh J, Norbert T, Sjorgren S, et al. Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat Med 1995;1:1029–34. [DOI] [PubMed] [Google Scholar]
- 12.Kovach JS, Hartmann A, Blaszyk H, et al. Mutation detection by highly sensitive methods indicates that p53 gene mutations in breast cancer can have important prognostic values. Proc Natl Acad Sci U S A 1996;93:1093–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elledge RM, Fuqua SAW, Clark GM, et al. Prognostic significance of p53 gene alterations in node-negative breast cancer. Breast Cancer Res Treat 1993;26:225–35. [DOI] [PubMed] [Google Scholar]
- 14.Beck T, Weller EE, Weikel W, et al. Usefulness of immunohistochemical staining for p53 in the prognosis of breast carcinomas: correlations with established prognosis parameters and with the proliferation marker, MIB-l. Gynecol Oncol 1995;57:96–104. [DOI] [PubMed] [Google Scholar]
- 15.Silvestrini R, Benini E, Veneroni S, et al. P53 and bcl-2 expression correlates with clinical outcome in a series of node-positive breast cancer patients. J Clin Oncol 1996;14:1604–10. [DOI] [PubMed] [Google Scholar]
- 16.Hori M, Nogami T, Itabashi M, et al. Expression of Bcl-2 in human breast cancer: correlation between hormone receptor status, p53 protein accumulation and DNA strand breaks associated with apoptosis. Pathol Int 1997;47:757–62. [DOI] [PubMed] [Google Scholar]
- 17.Seshadri R, Raymond WA, Leong AS, et al. Vimentin expression is not associated with poor prognosis in breast cancer. Int J Cancer 1996;67:353–6. [DOI] [PubMed] [Google Scholar]
- 18.Thomas PA, Kirschmann DA, Cerhan JR, et al. Association between keratin and vimentin expression, malignant phenotype, and survival in postmenopausal breast cancer patients. Clin Cancer Res 1999;5:2698–703. [PubMed] [Google Scholar]
- 19.Stampfer MR, Yaswen P. Culture systems for study of human mammary epithelial cell proliferation, differentiation and transformation. Cancer Surv 1993;18:7–34. [PubMed] [Google Scholar]
- 20.Taylor-Papadimitiou J, Stampfer M, Bartek J, et al. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: relation to in vivo phenotypes and influence of medium. J Cell Sci 1989;94:403–13. [DOI] [PubMed] [Google Scholar]
- 21.Kao CY, Nomata K, Oakley CS, et al. Two types of normal human breast epithelial cells derived from reduction mammoplasty. Phenotypic characterization and response to SV40 transfection. Carcinogenesis 1995;16:531–8. [DOI] [PubMed] [Google Scholar]
- 22.Cattoretti G, Andreola S, Clemente C, et al. Vimentin and p53 expression on epidermal growth factor receptor-positive, oestrogen receptor-negative breast carcinomas. Br J Cancer 1988;57:353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domagala W, Markiewski M, Harezga B, et al. Prognostic significance of tumor cell proliferation rate as determined by the MIB-1 antibody in breast carcinoma: its relationship with vimentin and p53 protein. Clin Cancer Res 1996;2:147–54. [PubMed] [Google Scholar]
- 24.Seshadri R, McLeay WR, Horsfall DJ, et al. Prospective study of the prognostic significance of epidermal growth factor receptor in primary breast cancer. Int J Cancer 1996;69:23–31. [DOI] [PubMed] [Google Scholar]
- 25.Domagala W, Lasota J, Bartkowiak J, et al. Vimentin is preferentially expressed in human breast carcinomas with low estrogen receptor and high Ki-67 growth fraction. Am J Pathol 1990;136:219–27. [PMC free article] [PubMed] [Google Scholar]
- 26.Doglioni C, Dei Tos AP, Laurino L, et al. The prevalence of Bcl-2 immunoreactivity in breast carcinomas and its clinicopathological correlates, with particular reference to oestrogen receptor status. Virchows Arch 1994;424:47–51. [DOI] [PubMed] [Google Scholar]
- 27.Leek RD, Kaklamanis L, Pezzella F, et al. Bcl-2 in normal human breast and carcinoma, association with oestrogen receptor-positive, epidermal growth factor receptor-negative tumours and in situ cancer. Br J Cancer 1994;69:135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koutselini H, Markopoulos C, Lambropoulou S, et al. Relationship of epidermal growth factor receptor (EGFR), proliferating cell nuclear antigen (PCNA) and vimentin expression and various prognostic factors in breast cancer patients. Cytopathology 1995;6:14–21. [DOI] [PubMed] [Google Scholar]
- 29.Raymond WA, Leong AS. Vimentin: a new prognostic parameter in breast carcinoma? J Pathol 1989;158:107–14. [DOI] [PubMed] [Google Scholar]
- 30.Dabbs DJ. Correlation of morphology, proliferation indices, and oncogene activation in ductal breast carcinoma: nuclear grade, S-phase, proliferating cell nuclear antigen, p53, epidermal growth factor receptor, and c-erb-B-2. Mod Pathol 1995;8:637–42. [PubMed] [Google Scholar]
- 31.World Health Organisation. Histological typing of breast tumors, 2nd ed. International histological classification of tumors, *2. Geneva: World Health Organisation, 1981.
- 32.Hermanek P, Sobin LH. UICC International Union Against Cancer. In: TNM classification of malignant tumors, 4th ed. Berlin: Springer-Verlag, 1987:93–9.
- 33.Megha T, Ferrari F, Lalinga AV, et al. Cellular kinetics and expression of bcl-2 and p53 in ductal carcinoma of the breast. Oncol Rep 2000;7:473–8. [DOI] [PubMed] [Google Scholar]
- 34.Leoncini L, Spina D, Close P, et al. Abortive mitoses and nuclear fragmentation in CD30+ large cells of Hodgkin's disease. Leuk Lymphoma 1996;22:119–24. [DOI] [PubMed] [Google Scholar]
- 35.van de Schepop HAM, de Jong S, van Diest PJ, et al. Counting of apoptotic cells: a methodological study in invasive breast cancer. Mol Pathol 1996;49:M214–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soong R, Robbins PD, Dix BR, et al. Concordance between p53 protein overexpression and gene mutation in a large series of common human carcinomas. Hum Pathol 1996;27:1050–5. [DOI] [PubMed] [Google Scholar]
- 37.Schmitt FC, Soares R, Cirnes L, et al. P53 in breast carcinomas: association between presence of mutation and immunohistochemical expression using a semiquantitative approach. Pathol Res Pract 1998;194:815–19. [DOI] [PubMed] [Google Scholar]
- 38.Bodis S, Siziopikou KK, Schmitt SJ, et al. Extensive apoptosis in ductal carcinoma in situ of the breast. Cancer 1996;77:1831–5. [DOI] [PubMed] [Google Scholar]
- 39.Kirsch DG, Kastan MB. Tumor-suppressor p53: implications for tumor development and prognosis. J Clin Oncol 1998;16:3158–68. [DOI] [PubMed] [Google Scholar]
- 40.Sierra A, Castelsagué X, Coll T, et al. Expression of death-related genes and their relationship to loss of apoptosis in T l ductal breast carcinomas. Int J Cancer 1998;79:103–10. [DOI] [PubMed] [Google Scholar]
- 41.Mommers E, van Diest PJ, Leonhart AM, et al. Balance of cell proliferation and apoptosis in breast carcinogenesis. Breast Cancer Res 1999;58:163–9. [DOI] [PubMed] [Google Scholar]
- 42.Ceraline J, Deplanque G, Duclos B, et al. Inactivation of p53 in normal human cells increases G2/M arrest and sensitivity to DNA-damaging agents. Int J Cancer 1998;75:432–8. [DOI] [PubMed] [Google Scholar]
- 43.Glinsky GV, Glinsky W, Ivanova AB, et al. Apoptosis and metastasis: increased apoptosis resistance of metastatic cancer cells is associated with the profound deficiency of apoptosis execution mechanisms. Cancer Lett 1997;115:185–93. [DOI] [PubMed] [Google Scholar]
- 44.de Jong JS, van Diest PJ, Michaelides RJAM, et al. Correlation of cyclin Dl and Rb gene expression with apoptosis in invasive breast cancer. Mol Pathol 1998;51:30–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuende E, Ales-Martinez A-F, Ding L, et al. Programmed cell death by bcl-2-dependent and independent mechanisms in B-lymphomas cells. EMBO J 1993;12:1555–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wynford-Thomas D. Proliferative life-span checkpoints: cell-type specificity and influence on tumour biology. Eur J Cancer 1997;33:716–26. [DOI] [PubMed] [Google Scholar]
- 47.Wynford-Thomas D, Jones CJ, Wyllie FS. The tumor suppressor gene p53 as a regulator of proliferative life-span and tumor progression. Biol Signals 1996;5:139–53. [DOI] [PubMed] [Google Scholar]