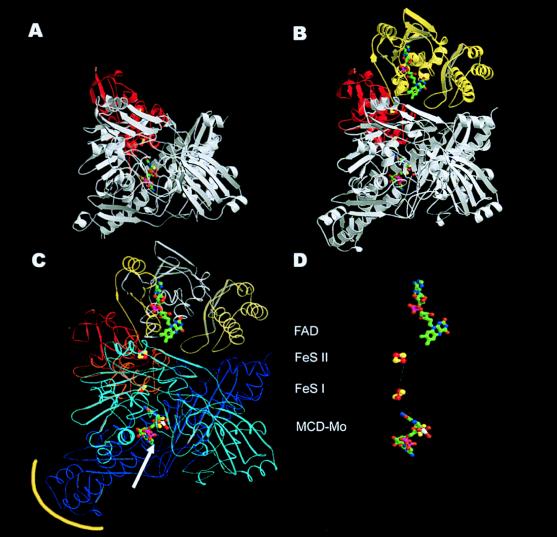
Figure 2.
Comparison of the CO dehydrogenase monomer (B) with the monomeric Mop structure from Desulfovibrio gigas (12) (A). Domain structure (C) and cofactors (D) of the CO dehydrogenase monomer. The ribbon presentation shows the general similarity of the two monomers for the MCD–molybdenum cofactor-containing subunit/domain (white) and the iron-sulfur containing subunit/domain (red) in both enzymes. The FAD-containing M subunit (yellow) is absent in the Mop protein. Whereas the S subunits are very similar in both proteins, the L subunit differs in a few areas. This applies mainly to the substrate channel of Mop (white arrow), which is closed by an additional loop in CO dehydrogenase and to the first part of the C-terminal domain (yellow encircled), which creates the dimer interface. (C) The three subunits of CO dehydrogenase may be subdivided into domains (the L subunit in a cyan N-terminal domain and a blue C-terminal domain). The S subunit has the iron-sulfur clusters bond in the N-terminal domain drawn in red and in the C-terminal domain drawn in orange. The three domains of the M subunit are yellow, white, and cream, sequentially, from N to C terminus. (D) The shortest connection between the cofactors is marked by a white line, starting from the molybdo-oxo group (MCD–Mo) at the active site via the proximal C-terminal [2Fe–2S] cluster (FeS I) (14.6 Å distance from the Mo atom to the closest iron atom) and the distal N-terminal [2Fe–2S] cluster (FeS II) (12.4 Å distance between the closest iron atoms), ending at the FAD (8.7 Å distance between C7 of FAD and the closest iron atom).