Abstract
Aim: The biology of colorectal hyperplastic polyps is of considerable relevance, because recent evidence suggests that under certain circumstances hyperplastic polyps may be precursors of neoplasms. The aim of this study was to assess and compare the clinical and molecular characteristics of hyperplastic polyps and neoplastic lesions removed from patients without the hyperplastic polyposis syndrome.
Methods: One hundred and twenty six patients were identified through a series of genetic epidemiological studies. Each patient had at least one neoplastic lesion and one hyperplastic polyp; there was a total of 147 hyperplastic polyps. All lesions were evaluated for K-ras mutations, loss of heterozygosity (LOH) of the adenomatous polyposis coli (APC) gene, and microsatellite instability.
Results: K-ras mutation was detected in 15 (10%) hyperplastic polyps, all from the rectosigmoid colon. No hyperplastic polyp had APC LOH or microsatellite instability. Patients with adenomas or carcinomas showing K-ras mutations were not more likely to have hyperplastic polyps with K-ras mutations. The average number of adenomas did not differ between those patients with hyperplastic polyps with K-ras mutations and those without K-ras mutations. There was no association between the hyperplastic polyp and the adenoma regarding the colon segments from which the two lesions were removed.
Conclusions: The sporadic hyperplastic polyp is a lesion with limited molecular change and no relation to patients’ neoplastic lesions.
Keywords: hyperplastic polyps, colonic adenomas, K-ras mutations, microsatellite instability, loss of heterozygosity
The hyperplastic polyp found in the colorectum is a growth with no phenotypic or morphological neoplastic features.1 It is defined by elongated crypts showing increased branching. There is a serrated pattern to the luminal surface. The biological importance of these polyps remains unclear, but hyperplastic polyps have received renewed interest in the past few years. There is increasing evidence that the hyperplastic polyp may under certain circumstances be a precursor for neoplasia.1
Recent reports have shown that, in common with adenomatous polyps, hyperplastic polyps may have K-ras mutations,2,3 DNA microsatellite instability,4–6 mutations of transforming growth factor βR11,5 rearrangements involving chromosome 1p,6,7 in addition to hypomethylation and CpG island methylation.8,9 However, much of this information is based upon analyses of hyperplastic polyps removed from patients with the hyperplastic polyposis syndrome (HPS), a rare condition involving dozens of hyperplastic polyps scattered throughout the colon. The true sporadic hyperplastic polyp occurs with much greater frequency. Published reports on hyperplastic polyps have each evaluated just 24 or fewer sporadic hyperplastic polyps.2,3,5–7,9–13
“There is increasing evidence that the hyperplastic polyp may under certain circumstances be a precursor for neoplasia”
The aim of our study was to examine the clinical and molecular characteristics of a large series of hyperplastic polyps from patients without HPS but with known neoplastic lesions, and to compare the findings between the hyperplastic polyps and the neoplastic lesions.
MATERIALS AND METHODS
Initial ascertainment of patients was through the endoscopy suite of a large, suburban community hospital and through hospital pathology records of outpatients and inpatients. The patients had all been evaluated by a group of 12 participating colonoscopists. Family and personal history were obtained by telephone interview. All patients had at least one adenoma or carcinoma. All hyperplastic polyps were recorded and studied simultaneously with the neoplastic lesions and adjacent normal mucosa. Patients with a family history or personal history suggestive of familial adenomatous polyposis, hyperplastic polyposis syndrome, or hereditary non-polyposis colorectal carcinoma (HNPCC) were excluded. All tissue samples were numerically coded. Haematoxylin and eosin stained slides of all study adenomas and hyperplastic polyps were histologically reviewed by one of two staff pathologists. There were no sessile serrated polyps included in our study group. Right sided lesions were those removed from the caecum, ascending, and transverse colonic segments. Left sided lesions were those removed from the descending, sigmoid, and rectal segments. The study design was reviewed and approved by the hospital institutional review board and, recently, its privacy subcommittee.
All specimens were formalin fixed and paraffin wax embedded, and were processed for DNA extraction and purification as reported previously.14 Loss of heterozygosity (LOH) of the adenomatous polyposis coli (APC) gene was determined through the polymerase chain reaction (PCR) amplification of a CA repeat marker within the D5S346 locus of the DP1 gene using the fluorescent labelled primer set: 5′–ACTCACTCTAGTGATAAATCGGG-3′ (sense) and 5′–AGCAGATAAGACAAGTATTACTAGTT-3 (antisense).15 Reactions were run on a PE 9700 thermal cycler (PE Applied Biosystems, Foster City, California, USA) under the following conditions: an initial denaturation for six minutes at 94°C, then 36 cycles of denaturation for 30 seconds at 94°, annealing for 30 seconds at 55°C, and elongation for one minute at 72°C, with a final 30 minute extension at 72°C.
Single stranded conformational polymorphism (SSCP) analysis was used to screen for mutations within the K-ras oncogene. The codon 12/13 region of exon 1 in the K-ras oncogene were amplified using the primer set 5′-CCTGCTGAAAATGACTGAAT-3′ (sense) and 5′-TGTTGGATCATATTCGTCCA-3′ (antisense). Samples were amplified and analysed as reported previously.14 Samples showing K-ras mutation bands by SSCP were sequenced to verify the point mutation. PCR based DyeDeoxy Terminator sequencing was performed using an ABI Prism 377 DNA sequencing system (Perkin Elmer, Foster City, California, USA) (fig 1).
Figure 1.
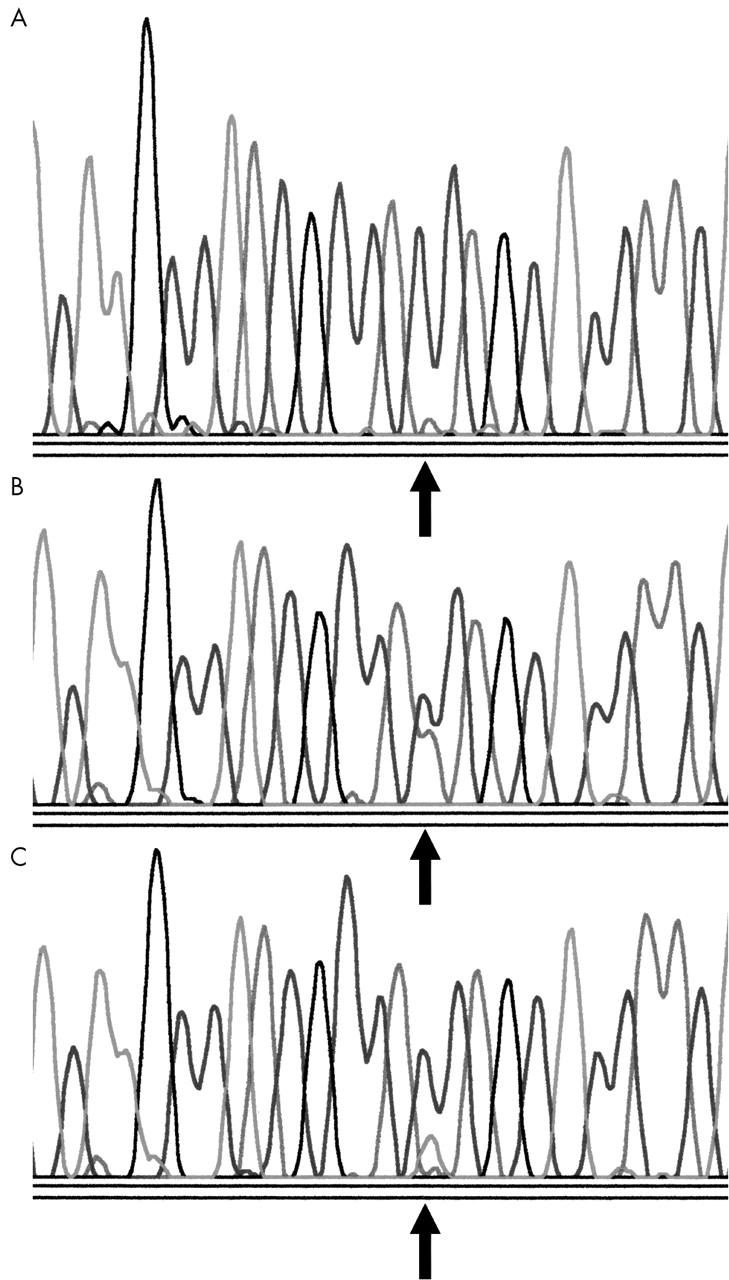
DNA sequence analysis of the K-ras gene. (A) Segment of the gene inclusive of codons 12 and 13. The arrow indicates the wild-type sequence at codon 12. (B) The arrow indicates a transversion of guanine to thymine in the second position of codon 12. (C) The arrow indicates a transition of guanine to adenine in the second position of codon 12.
Lesions were evaluated for microsatellite instability using a panel of three microsatellite markers, the mononucleotide repeat BAT-26 and the dinucleotide repeats D5S346 and D18S58. PCR primers for each marker were used to amplify DNA from normal and abnormal tissue. The amplified products were assayed side by side on an electrophoresis gel. Microsatellite instability for a given primer set was defined as the appearance of one or more new PCR products either larger or smaller than that produced by the DNA derived from normal tissue.
STATISTICAL METHODS
Mean and standard deviations were derived for patient age when the hyperplastic polyp was diagnosed, the number of adenomas for each patient, and the number of hyperplastic polyps for each patient. Fisher’s exact test was used to assess whether there were associations between the K-ras mutation status of a patient’s adenomas and the patient’s hyperplastic polyp, and the concordance between the location of the hyperplastic polyp and the adenoma. Odds ratios (OR) and 95% confidence intervals (CI) were derived as a measure of the strength of the association. The t test was used to assess whether the mean number of adenomas differed for patients with or without one or more hyperplastic polyp with a K-ras mutation.
RESULTS
Patient characteristics
From 1995 to 2002, 1150 patients were identified who had undergone one or more colonoscopy, had paraffin wax blocks of neoplastic colorectal tissue available through the pathology department, and met the criteria for one of several sequential genetic epidemiological studies. One hundred and twenty six of these patients were found to have had hyperplastic polyps. There were 42 women and 84 men. The mean (SD) age at the time of removal of the first hyperplastic polyp was 63.7 (10.8) years. Each patient had an average of 1.17 (0.47) hyperplastic polyps, and in total there were 147 polyps. One hundred and nine patients (87%) had only one hyperplastic polyp, whereas 14 patients (11%) had two. Only three patients (2%) had more than two hyperplastic polyps: two patients each had three and one had four hyperplastic polyps (table 1). Of the 126 patients, 89 patients (71%) had only a left sided hyperplastic polyp, whereas 32 patients (25%) had only a right sided hyperplastic polyp. Four patients (3%) had hyperplastic polyps on both sides of the colon, and one patient had one hyperplastic polyp of unknown location (table 1).
Table 1.
Patient demographics and hyperplastic polyp characteristics
| Patients | |
| Total patients | 126 |
| Sex | |
| Female | 42 (33%) |
| Male | 84 (67%) |
| Mean (SD) age at first HP | 63.7 years (10.8) |
| No of HPs/patient | |
| 1 | 109 (87%) |
| 2 | 14 (11%) |
| 3 | 2 (1.6%) |
| 4 | 1 (0.8%) |
| Mean (SD) number HP/patient | 1.17 (0.5) |
| HP location by patient | |
| Left side only | 89 (71%) |
| Right side only | 32 (25%) |
| Both sides | 4 (3%) |
| Unknown | 1 (0.8%) |
| Hyperplastic polyps | |
| Total number | 147 |
| Left colon | 109 (74%) |
| Right colon | 37 (25%) |
| Unknown site | 1 (0.7%) |
| Number with K-ras mutation | 15 of 143 assayed (10%) |
| Location of HPs with K-ras mutation | |
| Rectum | 5 |
| Sigmoid | 10 |
| Size of HP | |
| 15 HPs with K-ras mutation | 3.2 mm |
| 128 HPs without K-ras mutation | 3.25 mm |
| Number with APC LOH | 0 of 133 assayed |
| Number with MSI | 0 of 55 assayed |
| HP in same segment | |
| As an adenoma* | |
| Yes | 68 (48%) |
| No | 73 (52%) |
*1 patient had HP of unknown location, 4 had carcinoma and no adenoma, 1 had small bowel carcinoma.
APC, adenomatous polyposis coli; HP, hyperplastic polyp; LOH, loss of heterozygosity; MSI, microsatellite instability.
Adenomas
One hundred and twenty two patients had a mean (SD) of 3.2 (3.0) adenomas in addition to their hyperplastic polyps. Forty three patients (35%) had only one adenoma, whereas 28 patients (23%) had two adenomas, and 51 patients (42%) had three or more adenomas. Seventy (57%) of the 122 patients with adenomas had only tubular adenomas, whereas the other 52 patients (43%) had adenomas with some villous component (tubulovillous or villous) (table 2). Of the 122 patients with adenomas, 46 patients (38%) had left sided adenomas only, 24 patients (20%) had right sided adenomas only, and 52 patients (43%) had adenomas on both sides of the colon. Sixty nine patients (57%) had had an adenoma before the detection of their first hyperplastic polyp, with an average interval of 4.3 years (range, 1–19). Thirty two patients (26%) had an adenoma after their last hyperplastic polyp, with an average interval of 4.7 years (range, 1–15).
Table 2.
Neoplastic lesions in patients with hyperplastic polyps
| Patient characteristic | Patients with adenomas N (%) | Patients with carcinomas N (%) |
| No of patients with neoplastic lesions | 122 | 26 |
| Mean (SD) no of adenomas/patient | 3.2 (3) | N/A |
| No of patients with only tubular adenomas | 70 (57%) | N/A |
| No of patients with at least one tubulovillous or villous lesion | 52 (43%) | N/A |
| No of patients with at least one lesion with K-ras mutation | 42 (34%) | 8 of 23 (35%) |
| No of patients with K-ras mutation in both neoplastic lesion and HP | 3 (2%) | 0 |
| No of patients with at least one neoplastic lesion with APC LOH | 42 of 112 (38%) | 3 of 21 (14.3%) |
*APC, adenomatous polyposis coli gene; HP, hyperplastic polyp; LOH, loss of heterozygosity.
Of the 122 patients with adenomas, 42 patients (34%) had at least one adenoma with a K-ras mutation. However, of these 42 patients, only three had a hyperplastic polyp with a K-ras mutation, whereas 39 had a hyperplastic polyp with no Ki-ras mutation (table 2). In contrast, 80 patients had adenomas without a K-ras mutation, and 11 of these patients also had a K-ras mutated hyperplastic polyp. Thus, patients with hyperplastic polyps with a K-ras mutation were not more likely to have adenomas with K-ras mutations (OR, 0.48; 95% CI, 0.08 to 1.99). One hundred and twelve patients had at least one adenoma assayed for APC LOH, and 42 of them (38%) had at least one adenoma with APC LOH (table 2).
Carcinomas
Of the 126 patients, 26 had a carcinoma. Four patients had only a carcinoma and 22 had both a carcinoma and one or more adenomas. Sixteen carcinomas were in the left colon, nine were in the right colon, and one was located in the small bowel. Twenty three of the 26 carcinomas were assayed with respect to the K-ras gene and eight were found to have a K-ras mutation (table 2). None of the eight patients with K-ras mutated colorectal carcinoma had a hyperplastic polyp with a K-ras mutation. Of the four patients with only a carcinoma and no adenoma, one carcinoma was K-ras mutated; all four had a hyperplastic polyp with no K-ras mutation. APC LOH was detected in three of 21 carcinomas assayed. Among the 21 patients with only left sided hyperplastic polyps and a colorectal carcinoma, 13 had a left sided carcinoma and seven had a right sided carcinoma, whereas one patient had a small bowel carcinoma.
Hyperplastic polyps
In total, 147 hyperplastic polyps were removed from the 126 patients. One hundred and nine hyperplastic polyps (74%) were found in the left colon, 37 (25%) were removed from the right colon, and one was from an unknown location (table 1). The location by segment was: caecum, eight; ascending colon, 12; transverse colon, 17; descending colon, 15; sigmoid colon, 64; and rectum, 30. One hundred and forty three of the 147 hyperplastic polyps (97%) were assayed for Ki-ras mutation and in 15 (10%) a mutation was detected. These 15 hyperplastic polyps were removed from 14 patients, and they had been located in the rectum in five patients and in the sigmoid colon in 10 patients (table 1). At least one hyperplastic polyp with a K-ras mutation occurred in 7% of all women and 13% of all men, percentages that are not significantly different (p < 0.38; OR, 0.5; 95% CI, 0.1 to 2.0). Fourteen of the 15 mutations occurred within codon 12, namely: nine transitions GGT to GAT, four transversions GGT to GTT, one transversion GGT to GCT; in addition, there was one transversion in codon 13: GCC to GAC.
Of the fourteen patients who had two hyperplastic polyps, 11 patients had no K-ras mutation in either, one patient had a K-ras mutation in both, and two patients had one hyperplastic polyp with a K-ras mutation and one without. Of the two patients who had three hyperplastic polyps, one patient had no K-ras mutations, and the other patient had only one hyperplastic polyp with a K-ras mutation. The one patient with four hyperplastic polyps had three without a K-ras mutation and one with a K-ras mutation.
The 14 patients with K-ras mutated hyperplastic polyps had an average of 4.1 adenomas, compared with an average of 3.1 adenomas for the 108 patients with hyperplastic polyps that were K-ras normal, a difference that is not significant (p < 0.23).
We sized each hyperplastic polyp based upon the largest of the three dimensions measured by the pathologist. The mean size of the 15 hyperplastic polyps with a K-ras mutation was 3.2 mm. The mean size for the 128 hyperplastic polyps without a k-ras mutation was 3.25 mm (no size was recorded for four), a difference that is not significant (p < 0.9) (table 1).
One hundred and thirty three hyperplastic polyps were assayed for APC LOH, and none of them revealed LOH for the APC gene. The 14 hyperplastic polyps not assayed for APC LOH were from all segments of the colon. In total, 55 hyperplastic polyps were assessed for microsatellite instability: 32 of the 37 hyperplastic polyps from the right colon and 23 of the 109 hyperplastic polyps from the left colon (including all hyperplastic polyps found to have a Ki-ras mutation). These 55 hyperplastic polyps were all microsatellite stable (table 1).
One hundred and forty two hyperplastic polyps had been removed from colons that also yielded at least one adenoma. There was no association between the hyperplastic polyp and the adenoma regarding the colon segment or the side of the colon from which the two lesions were removed. Of these 142 hyperplastic polyps, 68 (48%) had been removed from the same segment as the adenoma, and 73 (52%) had come from a different segment than the adenoma (the location of one hyperplastic polyp was unknown) (table 1). In total, 90 patients had hyperplastic polyps on the left side, and 71 of them had adenomas also on the left side, whereas 19 patients had adenomas only on the right side of the colon. Of the 31 patients with hyperplastic polyps only on the right side of the colon, 26 patients had adenomas on the left side and five patients had adenomas only on the right side. There was no relation between the location of hyperplastic polyps and adenomas (OR, 0.72; 95% CI, 0.19 to 2.27).
In total, 25 patients had a carcinoma of the colon and one had a small bowel carcinoma. Twenty one patients had hyperplastic polyps in the left colon, 14 of whom had a carcinoma in the left side and seven of whom had a carcinoma in the right side. Four patients had hyperplastic polyps only on the right side of the colon, two of whom had a carcinoma in the left side, whereas two had a carcinoma in the right side.
We also identified six additional patients from our entire cohort who had hyperplastic polyps but no colorectal neoplasm (data not shown). These six patients had a total of nine hyperplastic polyps, three of which were K-ras mutated, two from the sigmoid colon and one from the rectum. None of the hyperplastic polyps had APC LOH, and the three with a K-ras mutation were microsatellite stable. Of note, one of the six patients had three hyperplastic polyps: one removed from the ascending colon, and two years later, one removed from the rectum and one from the transverse colon. Only the hyperplastic polyp from the rectum was K-ras mutated.
Histological review
As a separate evaluation, an independent pathologist with recognised expertise in gastroenterological pathology, who had not been involved with the activities of our study, re-reviewed a subset of haematoxylin and eosin stained slides. The 15 hyperplastic polyps with K-ras mutation were admixed with 15 other hyperplastic polyps without a K-ras mutation. Our expert pathologist reviewed all 30 cases blindly. He could detect no specific histological features unique to those cases with K-ras mutations (fig 2).
Figure 2.
A sigmoid hyperplastic polyp with a mutation in the K-ras gene: GGT to GAT in codon 12.
DISCUSSION
True hyperplastic polyps, also called metaplastic polyps, have a serrated pattern as their most distinctive histological feature. Hyperplastic polyps are a localised epithelial hyperplasia, yet the overall normal cellular organisation is maintained.16 Two types of mixed polyps are also recognised and frequently confused with the true hyperplastic polyp. The first has been described as a “collision” of a typical hyperplastic polyp and a tubular adenoma, showing features of both. The second, called a serrated adenoma, is dysplastic throughout, but it shows epithelial serration similar to that of a hyperplastic polyp. We have limited our study to only the true, sporadic, hyperplastic polyp.
Several reports of molecular genetic changes in hyperplastic polyps included only a limited number of incidental hyperplastic polyps, along with numerous other lesions.2,3,5–7,9–13 These 10 studies have formed the basis for numerous conclusions regarding hyperplastic polyps, yet they represent studies on just 127 isolated hyperplastic polyps, whereas 1017 other colorectal non-cancerous lesions were also analysed. From these reports, 17 of 66 hyperplastic polyps had a K-ras mutation. APC LOH or APC mutations were not detected in 50 hyperplastic polyps, and microsatellite instability was found in just seven of 28 such lesions. In contrast, a recent study evaluated 90 hyperplastic polyps removed from 40 patients known to have HNPCC and found that all hyperplastic polyps showed full expression of the three mismatch repair proteins analysed, suggesting that mismatch repair dysfunction in hyperplastic polyps of HNPCC is very rare and probably not a key step in the carcinogenic pathway for HNPCC.17 Furthermore, in carcinoma, the methylation pathway results in dysfunction of genes responsible for DNA repair, which in turn leads to unrepaired replication errors and microsatellite instability. Although we did not directly assay for methylation, we did assay for microsatellite instability in many of our hyperplastic polyps but found none.
We did not survey a consecutive series of individuals undergoing colonoscopy and thus do not provide a prevalence of hyperplastic polyps in our general population. However, because all the patients who we analysed had neoplastic lesions, it is possible that their colonic mucosa would be more likely to give rise to hyperplastic polyps of pathological relevance, if indeed hyperplastic polyps are integral to adenoma or carcinoma development.
“Despite the removal of the hyperplastic polyp, 26% of patients developed subsequent adenomas”
The sex ratio of our patients was 2 : 1 (male : female). A similar ratio of 2.3 was reported previously for a large cohort of individuals undergoing colonoscopy during which one or more adenoma was found.18 The hyperplastic polyps of our patients were clearly incidental, because the average was just 1.17 hyperplastic polyps/patient, despite many individuals having had colonoscopies repeatedly over many years. Indeed, the average number of adenomas for the 122 patients having had adenomas was 3.2, indicating that the development of adenomas for this cohort was more common than the development of hyperplastic polyps.
The randomness of the development of hyperplastic polyps is perhaps indicated by the findings that 57% of those with adenomas had had adenomas removed at a time when they had no hyperplastic polyps, only to develop one subsequently. Furthermore, despite the removal of the hyperplastic polyp, 26% of patients developed subsequent adenomas.
We found that 74% of the hyperplastic polyps were in the left colon, consistent with previous studies. Of interest, however, was the finding that all 14 hyperplastic polyps with a mutation in the K-ras gene and known location were removed from the rectum and sigmoid colon. These hyperplastic polyps made up only a small fraction (14%) of all left sided hyperplastic polyps. Fourteen hyperplastic polyps had a K-ras mutation in codon 12 and one had a mutation in codon 13. The types of K-ras mutations were similar to our previous findings in adenomas, with GTT and GAT representing most of the mutations.12
Take home messages.
Sporadic hyperplastic polyps show limited molecular changes and are not related to the patients’ neoplastic lesions
Mutations in the K-ras gene were found mainly in codon 12, and only in hyperplastic polyps from the distal left colon
Conclusions regarding the pathological importance of hyperplastic polyps found in other clinical settings, such as the hyperplastic polyposis syndrome, may not be applicable to true sporadic hyperplastic polyps
There were no identifying microscopic histopathological features of the hyperplastic polyps with a K-ras mutation compared with those without a K-ras mutation. Our data with regard to the patients’ accompanying lesions suggest that the K-ras mutations in the rectosigmoid hyperplastic polyps do not have major pathological importance. The mutation may represent a sensitivity of the mucosa from the left side of the colon to mutagenic influences. None of our hyperplastic polyps had APC LOH, and all lesions studied were microsatellite stable. Patients with K-ras mutations in one or more of their adenomas or in their carcinoma were not more likely to have had a hyperplastic polyp with a K-ras mutation. Furthermore, there was no association between the specific colon segment, or which side of the colon that the hyperplastic polyp was located and the location of the patient’s adenomas or carcinoma.
Our data from a large cohort of 126 patients with 147 hyperplastic polyps reveals the sporadic hyperplastic polyp to be a lesion with limited molecular changes and with no positional or temporal relation to patients’ neoplastic lesions. Mutations in the K-ras gene were primarily in codon 12, and they were found only in hyperplastic polyps from the distal left colon. Conclusions regarding the pathological importance of the hyperplastic polyp found in other clinical settings, such as the HPS, may not be applicable to the truly sporadic hyperplastic polyp.
Acknowledgments
The authors acknowledge the expert review of histological slides by Dr R R Rickert and the ongoing review of pathological material by Dr E Berman. This research was supported by the Harvey Nussbaum Research Foundation of Saint Barnabas Medical Center, the Walter and Louise Sutcliffe Foundation, and Cancer Research UK.
Abbreviations
APC, adenomatous polyposis coli
CI, confidence interval
HPS, hyperplastic polyposis syndrome
LOH, loss of heterozygosity
OR, odds ratio
PCR, polymerase chain reaction
SSCP, single stranded conformational polymorphism
REFERENCES
- 1.Jass JR. Hyperplastic polyps of the colorectum—innocent or guilty? Dis Colon Rectum 2001;44:163–6. [DOI] [PubMed] [Google Scholar]
- 2.Otori K , Oda Y, Sugiyama K, et al. High frequency of K-ras mutations in human colorectal hyperplastic polyps. Gut 1997;40:660–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jen J , Powell SM, Papadopoulos N, et al. Molecular determinants of dysplasia in colorectal lesions. Cancer Res 1994;54:5523–6. [PubMed] [Google Scholar]
- 4.Jeevaratnam P , Cottier DS, Browett PJ, et al. Familial giant hyperplastic polyposis predisposing to colorectal cancer: a new hereditary bowel cancer syndrome. J Pathol 1996;179:20–5. [DOI] [PubMed] [Google Scholar]
- 5.Konishi M , Kikuchi-Yanoshita R, Tanaka K, et al. Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology 1996;111:307–17. [DOI] [PubMed] [Google Scholar]
- 6.Lothe RA, Andersen SN, Hofstad B, et al. Deletion of 1p loci and microsatellite instability in colorectal polyps. Genes Chromosomes Cancer 1995;14:182–8. [DOI] [PubMed] [Google Scholar]
- 7.Bardi G , Pandis N, Fenger C, et al. Deletion of 1p36 as a primary chromosomal aberration in intestinal tumorigenesis. Cancer Res 1993;53:1895–8. [PubMed] [Google Scholar]
- 8.Bariol C , Suter C, Cheong K, et al. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol 2003;162:1361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan AO, Issa JJ, Morris JS, et al. Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol 2002;160:529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchida H , Ando H, Maruyama K, et al. Genetic alterations of mixed hyperplastic adenomatous polyps in the colon and rectum. Jpn J Cancer Res 1998;89:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iino H , Jass JR, Simms LA, et al. DNA microsatellite instability in hyperplastic polyps, serrated adenomas, and mixed polyps: a mild mutator pathway for colorectal cancer? J Clin Pathol 1999;52:5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogt F , Brien T, Brown CA, et al. Genetic alterations in serrated adenomas: comparison to conventional adenomas and hyperplastic polyps. Hum Pathol 2002;33:87–91. [DOI] [PubMed] [Google Scholar]
- 13.Odze RD, Brien T, Brown CA, et al. Molecular alterations in chronic ulcerative colitis-associated and sporadic hyperplastic polyps: a comparative analysis. Am J Gastroenterol 2002;97:1235–42. [DOI] [PubMed] [Google Scholar]
- 14.Zauber NP, Sabbath-Solitare M, Marotta SP, et al. Molecular changes in the Ki-ras and APC genes in colorectal adenomas and carcinomas arising in the same patient. J Pathol 2001;193:303–9. [DOI] [PubMed] [Google Scholar]
- 15.Spirio L , Nelson L, Joslyn G, et al. A CA repeat 30–70 kb downstream from the adenomatous polyposis coli (APC) gene. Nucleic Acids Res 1991;19:6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams GT. Metaplastic (hyperplastic) polyps of the large bowel: benign neoplasms after all? Gut 1997;40:691–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rijcken FEM, Sluis T, Hollema H, et al. Hyperplastic polyps in hereditary nonpolyposis colorectal cancer. Am J Gastroenterol 2003;98:2306–11. [DOI] [PubMed] [Google Scholar]
- 18.Winawer SJ, Zauber AG, Ho MN, et al. Prevention of colorectal cancer by colonoscopic polypectomy. N Engl J Med 1993;329:1977–81. [DOI] [PubMed] [Google Scholar]