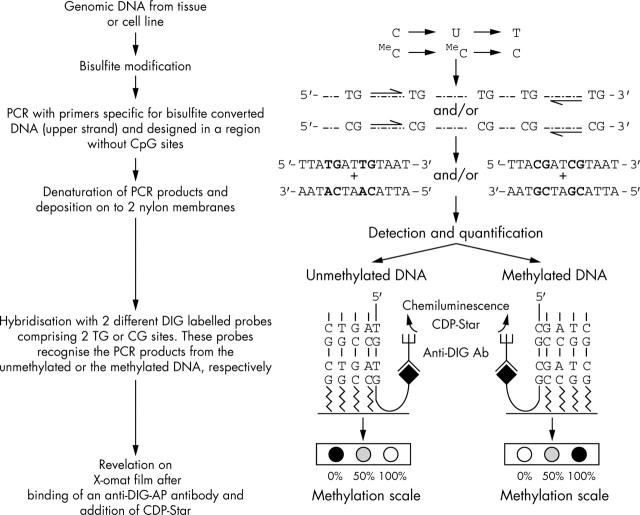
Figure 1.
Outline of the methylation sensitive dot blot assay. The method is based on bisulfite modification, polymerase chain reaction amplification using primers without CpG sites and specific to the modified strand, followed by a quantitative, non-radioactive dot blot readout. The use of two DIG labelled oligoprobes specific for either the methylated or the unmethylated DNA leads to different readout patterns: no methylation, full methylation, or a combination of unmethylated and methylated alleles.