Abstract
Background: Breast cancer is increasing in Indonesia and other developing countries. Germline mutations in the BRCA1/2 genes are most strongly associated with a high risk for breast cancer development. There have been no reports on BRCA1/2 gene mutations in the Indonesian population. Genetic research yielding insight into mutations affecting the Indonesian population can help in risk assessment of individual patients.
Aims: To screen the BRCA1/2 genes for mutations in early onset Indonesian breast cancer patients and their families with a new, simple, and sensitive BRCA1/2 mutation screening strategy based on denaturing gradient gel electrophoresis (DGGE) and targeted sequencing.
Methods: DNA was isolated from the blood of four Indonesian breast cancer patients from high risk families and seven family members, and the polymerase chain reaction was performed with specially designed primers throughout the BRCA1/2 coding sequences to produce fragments suitable for pooled DGGE analysis. The aberrantly migrating samples were reamplified and sequenced.
Results: Two mutations were found in exons 13 and 16 of BRCA1 and two mutations in exons 2 and 14 of BRCA2, which turned out to be established polymorphisms according to the Breast Cancer Information Core. In addition, a novel 6 bp deletion in exon 11, leading to a premature stop, was found in BRCA2.
Conclusion: Pooled DGGE and targeted sequencing revealed four BRCA1/2 polymorphisms and one novel BRCA2 mutation in a group of Indonesian patients at high risk of hereditary breast cancer. This illustrates that the proposed method is sensitive and particularly suited for screening unknown populations.
Keywords: BRCAA1, BRCA2, hereditary breast cancer, methodology, mutation detection
Breast cancer is the most common malignancy in women in the Western world, accounting for 32% of all female cancers, and is responsible for 18% of cancer deaths in women. The hereditary form of breast cancer constitutes about 5% of breast cancer cases overall.1 The first major gene responsible for hereditary breast cancer susceptibility, BRCA1, was mapped to 17q21 in 1990,2 and was cloned in 1994.3 A second gene involved in the hereditary breast cancer syndrome, BRCA2, was identified several months later.4 Both genes have long coding sequences and complex genomic structures: BRCA1 comprises 5592 bp, composed of 22 coding exons that encode 1863 amino acids,3 and BRCA2 comprises 10 443 bp, with 26 coding exons that encode 3418 amino acids.4 In addition to breast cancer, ovarian cancer,5 fallopian tube cancer,6 and primary peritoneal cancer are part of the BRCA1/2 cancer spectrum in women.
“By identifying the endemic Indonesian mutations, we hope to offer better risk assessment for women in the Indonesian population who are susceptible to the BRCA1/2 related hereditary cancers”
Over 80% of families with two or more cases of premenopausal breast cancer and two or more cases of ovarian cancer are believed to carry a germline BRCA1 or BRCA2 mutation.7,8 Comprehensive evaluation of breast cancer susceptibility genes can be used to provide women with information concerning their risk of developing cancer, and guide the decision on the necessity of preventive surgical measures.
It has been shown that ethnically different populations exhibit different germline mutation spectra in the BRCA1 and BRCA2 genes.9–11 There are no previous publications on BRCA1/2 mutation detection in the Indonesian population. By identifying the endemic Indonesian mutations, we hope to offer better risk assessment for women in the Indonesian population who are susceptible to the BRCA1/2 related hereditary cancers. There are no exact data on breast cancer incidence in Indonesia, but a study by Soeripto et al in 1982 (unpublished results) showed that the age standardised incidence rate in the Jogjakarta region was 6.17, ranking second after cervical cancer (7.69). One of the central themes in cancer risk assessment is the efficiency and accuracy of genetic screening methods. Currently, the ability to conduct large scale, population based studies is constrained by the lack of an accurate and inexpensive method for mutation detection. The large size of the BRCA1 and BRCA2 genes, and the scattered distribution of mutations throughout the genes, complicate the task of mutation detection and make rapid screening for mutations a major technical challenge. A technique that scans stretches of DNA for unknown mutations should be optimised to detect as close to 100% of the sequence alteration as possible. Here, we describe the results of the application of a recently developed rapid and sensitive method for the detection of BRCA1/2 mutations12 based on denaturing gradient gel electrophoresis (DGGE)13 and targeted sequencing on an Indonesian group of high risk patients. This method appears to be particularly suited for the analysis of unknown populations.
MATERIALS AND METHODS
Patients
From 1996 to 1998, four Indonesian families were selected from patient pools at three hospitals in Jogjakarta, Indonesia; three with hereditary breast cancer based on Moller’s criteria,14 and one with one early onset patient without a family history. Figure 1 shows the pedigrees of these four families. Information regarding potential risk factors for breast cancer was obtained through a structured face to face interview. Information on family history (up to the date of diagnosis of the patient’s breast cancer) was elicited by asking each patient to identify all first and second degree female blood relatives. For each identified relative, the interviewer then asked the year of birth, vital status, year of death (if applicable), history, and type of cancer (if any). The patients were interviewed for family history using standardised questionnaires and blood was taken after informed consent.
Figure 1.
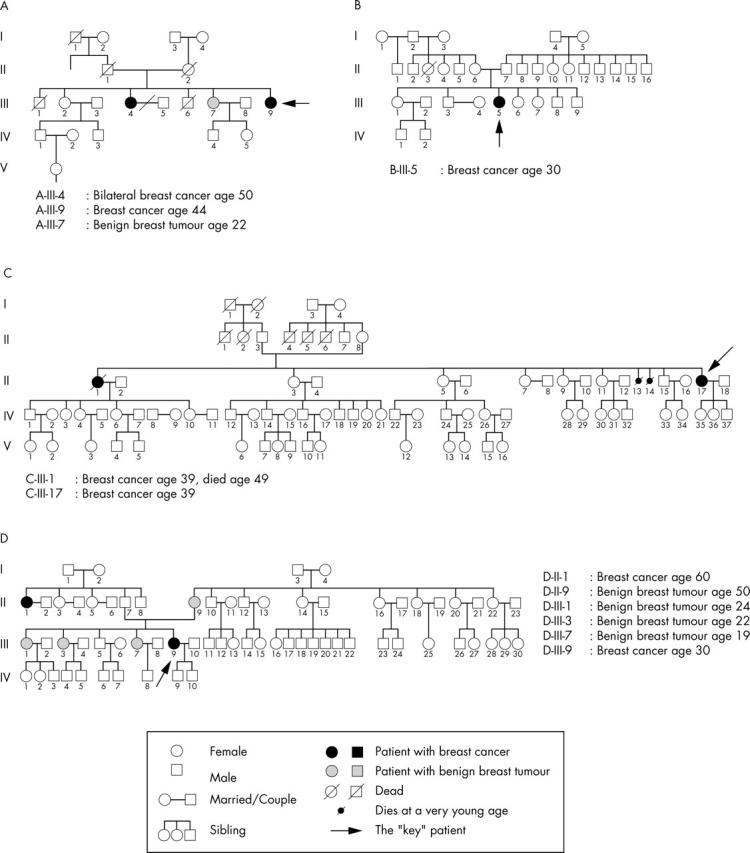
Pedigrees of four Indonesian families screened for BRCA1/2 mutations with a novel, rapid, and sensitive mutation detection method based on pooled denaturing gradient electrophoresis and targeted sequencing.
DNA amplification
DNA was isolated from peripheral blood using simple and rapid genomic DNA extraction. Two types of buffer—cell membrane lysis buffer, containing sucrose and triton X-100, and nuclear membrane lysis buffer, containing guanidine thiocyanate, sodium N-lauroyl sarcocinate, and β mercaptoethanol—were used. By using this method, only a small amount of peripheral blood (0.5 ml) was needed. The DNA was stored at 4°C.
Polymerase chain reaction (PCR) fragments suitable for DGGE analysis were designed based on the theoretical melting profile of each exon, as described previously.12 A 40 mer GC clamp was attached to the short oligonucleotide to obtain optimal melting profiles. Tables 1 and 2 provide the PCR primer sequences for BRCA1 and BRCA2, respectively.
Table 1.
Primer sequences used to amplify BRCA1 exon by exon for denaturing gradient gel electrophoresis12
| Fragment | Forward primer | Reverse primer | Size (bp) |
| Exon 2-1 | ATGATAAAATGAAGTTGTC* | ACACTCTAAGATTTTCTGC | 204 |
| Exon 2-2 | TTATCTGCTCTTCGCGTTG | CTTCCCTAGTATGTAAGGTC* | 202 |
| Exon 3 | GCGCGTTGAGCCTCATTTATTTTC | ACAAAAGCTAATAATGGAGC* | 185 |
| Exon 5 | GTATTCTTTCTACAAAAGG* | TCCAACCTAGCATGATTAC | 208 |
| Exon 6 | GGTTGATAATCACTTGCTG* | CACTTGAGTTGCATTCTTG | 223 |
| Exon 7 | ATACATAGGGTTTCTCTTG* | AGAAGAAAACAAATGGTTT | 293 |
| Exon 8 | TTGCTTGACTGTTCTTTACC* | ACTTAAAAAACCTGAGACC | 209 |
| Exon 9 | CCCTTTTAATTAAGAAAAC* | ACTAAATAGGAAAATACCAG | 191 |
| Exon 10 | CATTTGACAGTTCTGCATAC* | TTCAGTGCCTGTTAAGTTG | 217 |
| Exon 11-1 | ATGACAATTCAGTTTTTGAG* | TATTACTGGGTTGATGATG | 147 |
| Exon 11-2 | AGCTGCTTGTGAATTTTCTG* | ATAAACTGCTGTTCTCATGC | 243 |
| Exon 11-3 | TTTTTACAAATACTCATGCCAGCTC* | TAGGATTCTCTGAGCATGGC | 313 |
| Exon 11-4 | TTTTTTTTTTTGTGTGAGAGAAAAGAATGG* | CATCTACCTCATTTAGAACG | 272 |
| Exon 11-5 | GAATCAAATGCCAAAGTAGC* | GGACGCTCTTGTATTATCTG | 329 |
| Exon 11-6 | ATTATAGGAGCATTTGTTAC | TTTTCGAGTGATTCTATTGG* | 324 |
| Exon 11-7 | CAAAAGGTGATTCTATTCAG* | ATAAGGTGGGCTTAGATTTC | 275 |
| Exon 11-8 | CAGGCATATTCATGCGCTTG* | GAAAGTATCGCTGTCATGTC | 281 |
| Exon 11-9 | GAGTAACAAGCCAAATGAAC | GGGGTCTTCAGCATTATTAG* | 242 |
| Exon 11-10 | TTGTCAATCCTAGCCTTCC* | ATTAGTCCCTTGGGGTTTTC | 324 |
| Exon 11-11 | AATAAATGTGTGAGTCAGTG* | ACATTCCTCTTCTGCATTTC | 300 |
| Exon 11-12 | ATTCAAGGTTTCAAAGCGCC | GTATATTAACTGTCTGTAC* | 238 |
| Exon 11-13a | AGGAAGAAAATCAAGGAAAG* | TAATGAGTCCAGTTTCGTTG | 223 |
| Exon 11-13b | GCCAAATGTAGTATCAAAGG | CAGGTGACATTGAATGTTCC* | 248 |
| Exon 11-13c | AAAATCTGCTAGAGGAAAAC | TCATCACTGGAACCTATTTC* | 259 |
| Exon 11-13d | TAAAGAAGCCAGCTCAAGC | CTGAAATCAGATATGGAGAG* | 325 |
| Exon 11-13e | GCAAGAATGTCAAGAAGTAG* | CCATCATCTAACAGGTCATC | 181 |
| Exon 11-13f | AGTCATGCATCTCAGGTTTG* | ATAAGTTCTCTTCTGAGGAC | 280 |
| Exon 11-14 | CTTTCACCCATACACATTTG | TGCAGTCATTTAAGCTATTC* | 277 |
| Exon 11-15 | GAGTGTCTGTCTAAGAACAC* | TATTTGCAGTCAAGTCTTCC | 221 |
| Exon 11-16 | GTTTCTTCACAGTGCAGTG | AAATAGACTGGGGCAAACAC* | 296 |
| Exon 12 | GTCTGCTTTTACATCTGAACC* | AATGCAAAGGACACCACACAC | 221 |
| Exon 13 | GCGCGATTTCATTTTCTTGGTGCC | GGGAAGGAAAGAATTTTGC* | 305 |
| Exon 14 | TCAGAACAAAGCAGTAAAG* | AAGATGTCAGATACCACAG | 257 |
| Exon 15-1 | ATTGGTGGCGATGGTTTTC* | CTCCTCCACATCAACAACCT | 204 |
| Exon 15-2 | ACTACCCATCTCAAGAGGAG | AAATCAAAGTGTTTGTTCC* | 195 |
| Exon 16-1 | GACCAGAACTTTGTAATTC* | CCAGCAGTATCAGTAGTAT | 299 |
| Exon 16-2 | AAAGTTGCAGAATCTGCCC | TAAGTCTTAGTCATTAGGG* | 252 |
| Exon 17 | GTGCTAGAGGTAACTCATG* | CAGCAGATGCAAGGTATTC | 213 |
| Exon 18 | ACAGCACTTCCTGATTTG | TCTGAGGTGTTAAAGGGAG* | 222 |
| Exon 19 | TCTATCTCCGTGAAAAGAG | CTGGTTAGTTTGTAACATC* | 176 |
| Exon 20 | TGCTCCACTTCCATTGAAG* | TTTGTCAACTTGAGGGAGG | 220 |
| Exon 21 | CCTCTCTCCATTCCCCTG* | AAGGCTGGTGCTGGAACTC | 182 |
| Exon 22 | GCCTGGGTTAAGTATGCAG* | ATTGTGTCCTCCCTCTCTG | 210 |
*GC clamp sequence (CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCG) is attached to this site.
Table 2.
Primer sequences used to amplify BRCA2 exon by exon for denaturing gradient gel electrophoresis12
| Fragment | Forward primer | Reverse primer | Size (bp) |
| Exon 2 | TCCCTGTGTAAGTGCATT | CACTTTCTCGGTGTAATT* | 229 |
| Exon 3 | ACTAAGGTGGGATTTTTT* | CGCCCCCCCAGTCTACCATATTG | 350 |
| Exon 4 | CACTGAATTATTGTACTG | ATATGTAGGAAAATGTTTC* | 226 |
| Exon 5 | AAATAACCTAAGGGATTT | CATTTCTAGTATTCTAAG* | 170 |
| Exon 6 | CTTAACAATTTTCCCCTT | GCTATTGTCAAATTCTCA* | 169 |
| Exon 7 | GATCAGGGCATTTCTATA* | CGCCCCTCATCTGCTCTTTCTTG | 248 |
| Exon 8 | GTTTTTGCATTCTAGTGAT* | GTTAGCAATTTCAACAGTCT | 184 |
| Exon 9 | GAGAGTTTTTATACTAGTGA* | ACAGAGCAAGACTCCACCTC | 278 |
| Exon 10-0 | ATGTGCTTCTGTTTTATACT* | CGCCCGCCAAACTATCTTCTTCAGAGGT | 205 |
| Exon 10-1 | TTTTTTTTTTAGTATATGAAACAGTTGTAG* | CTTCTGATTTGCTACATTTG | 291 |
| Exon 10-2 | CTCATTTGTATCTGAAGTGG* | CGCCCTTTTGGTCACATGAAGAAAT | 255 |
| Exon 10-3 | GGAGCCCAGATGGCGAAAA* | CTGTTTCCTCATTTAATGGC | 234 |
| Exon 10-4 | GCGCCACGTATTTCTAGCCTACC | TTAAAGTTTGGATCAGTCAT* | 294 |
| Exon 10-5 | GAGAATCACCTAAAGAGACT* | GGGGGGCCAGCTTCCATTATCAAT | 210 |
| Exon 10-6 | GTTTGCTCACAGAAGGAGGA* | CCTGCATTCTTCAAAGCTAC | 256 |
| Exon 10-7 | AGCCACCACCACACAGAATT | CTTTCGGTATTTTTTTTCCT* | 171 |
| Exon 10-8 | GCTATACATGATGAAACATC* | GTACCTGAATCAGCATTTGC | 177 |
| Exon 10-9 | GCTTTTGAAGCACCACTTAC | GGAATCGTCATCTATAAAAC* | 138 |
| Exon 11-1 | CGCGTGAATGTGATTGATGGTAC | CTGTAGTTTTTCCTTATTAC* | 244 |
| Exon 11-2 | ATCTTGATTATAAAGAAGCA* | TGAATGTTGTACTGGGTGAC | 219 |
| Exon 11-3 | AAAAGAAGAGGTCTTGGCTG | GACTAGGTTTGACAGAACA* | 214 |
| Exon 11-4 | AGCACTCTTATTTTAACTCC | TTTTCAGGTGGCAACAGCTC* | 255 |
| Exon 11-5 | AATGTCAGACAAGCTCAAAG* | TTGGATTACTCTTCGATTTG | 245 |
| Exon 11-6 | TAGCATCACCTTCAAGAAAG* | CTGTGTCTCCATATAAAACC | 316 |
| Exon 11-7 | TGACTTGTGTAAACGAACCC | GTCCTGCCCATTTGTTCATG* | 286 |
| Exon 11-8 | GGACATCTCCTTGAATATAG* | TTCAACACAAGCTAAACTAG | 265 |
| Exon 11-9 | GAGCAAAATGTTCTTCAAAG* | AATTTCTGCCTTTTGGCTAG | 301 |
| Exon 11-10 | CCCCTCAGATGTTATTTTCC* | GATCAGCATCTCTGCATTCC | 271 |
| Exon 11-11 | AAGTGCCTGAAAACCAGATG | ATGAGCAGAATAAAAGCCCC* | 282 |
| Exon 11-12 | AGTGCTTCTGGTTATTTAAC | CTTACAGTTTTATCATTATG* | 285 |
| Exon 11-13 | GCAGAGGTACATCCAATAAG* | TTACTTGAATCACTGCCATC | 312 |
| Exon 11-14 | GAAGATAACAAAATATACTGC* | TTACCATGACATGCTTCTTG | 288 |
| Exon 11-15a | GAAGATTTGTCAGATTTAAC | TATGTCAGAATCTAATTCAG* | 310 |
| Exon 11-15B | ATCAGAAACCAGAAGAATTG* | TACCAACTGGGACACTTTCT | 184 |
| Exon 11-15C | CAGACATAGTTAAACACAAA | ATTTCACTAGTACCTTGCTC* | 265 |
| Exon 11-15D | AGGAATCTTTGGACAAAGTG | GTTTTGAGATTTTCAGTTTG* | 325 |
| Exon 11-15E | TTCTATTGAGACTGTGGTGC* | TGACCATCAAATATTCCTTC | 327 |
| Exon 11-15F | TGTGAGTCAGACTTCATTAC | GTTAGACATGCTACTGTTAC* | 250 |
| Exon 11-15G | GTACTATAGCTGAAAATGAC* | TCCTCAACGCAAATATCTTC | 319 |
| Exon 11-15H | AAAAGATGCAAATGCATACC | CCCTACCTCAAAATTATTAC* | 179 |
| Exon 11-15I | ATGCAGCCATTAAATTGTCC | TACTTTACTGAAACTGTCTG* | 197 |
| Exon 11-15J | CAGTGGTAAAATCGTTTGTG* | TCTTCACTCTGAATGTCAGC | 283 |
| Exon 11-15K | TTATGATGAATGTAGCACGC | CTTCCCTATACTACATTTAC* | 206 |
| Exon 11-15L | CACCTTGTGATGTTAGTTTGG | ACCACATTATATGAAAAGCC* | 362 |
| Exon 11-15M | GAAGAAAATACTGCTATACG* | ATCAAATTCCTCTAACACTC | 211 |
| Exon 11-15N | TACAGCAAGTGGAAAGCAAG | TTATTTTCTGAAGAACCACC* | 301 |
| Exon 11-16 | CTTCCTCGTGTTGATAAGAG* | GGCATGACTTGGCAGTTTAG | 472 |
| Exon 11-17 | GGAAGATGATGAACTGACAG | GTGATTGGCAACACGAAAGG* | 214 |
| Exon 12 | GACTTTTGAGAAATAAAACTG* | GATCCACCTGAGGTCAGAAT | 278 |
| Exon 13 | CGCCCGTAATATAAAATAATGGTTTCC* | AAACGAGACTTTTCTCATAC | 196 |
| Exon 14-1 | CGCCCGCCGTGTACTAGTCAATAAACTT | AGCAGAAACTTGATAAAATG* | 244 |
| Exon 14-2 | ATCTTCAAGCAATTTAGCAG* | TACTATCATCAGAGCCATGT | 277 |
| Exon 14-3 | AAAACAGACAAAAGCAAAAC* | CATCACACAATTGTCATAC | 225 |
| Exon 15-1 | GGTGTGCTTTTTAAATTC* | CTTCTTAATTCGCATATCCTG | 151 |
| Exon 15-2 | ATTACAAGTCTTCAGAATGCC* | GCGGGAAAAGCCATCAGTATTGTAG | 254 |
| Exon 16 | GTGTGATACATGTTTACTT* | GGTAGAGGGAATACATAAA | 358 |
| Exon 17 | CAGTATCATCCTATGTGGTT* | CTGCCGTATATGATTACGTA | 303 |
| Exon 18-1 | CTCAGTTATTCAGTGACTTG* | GATCTAACTGGGCCTTAACA | 354 |
| Exon 18-2 | TTGAACTTACAGATGGGTGG | CTGATTTTTACCAAGAGTGC* | 248 |
| Exon 19 | CGCCCGCCATATTTATTAATTTGTCC | TATGGTAAGTTTCAAGAATAC* | 252 |
| Exon 20 | TATGTGACTTTTTTGGTGTG | CTCTAAGACTTTGTTCTCAT* | 281 |
| Exon 21-1 | GTGAATTAATAATCCTTTTG* | GCACGAACTTGCTGTCTTGT | 127 |
| Exon 21-2 | CATATTTACCATCACGTGCA | GCCTCATTATATGTCCTCTT* | 177 |
| Exon 22-1 | CTAGTTACAATAGATGGAAC* | TGTGACATCCCTTGATAAAC | 254 |
| Exon 22-2 | TGGAATCTGCTGAACAAAAG | CTGATAAAAACAAAGCATTTAC* | 169 |
| Exon 23-1 | GATAATCACTTCTTCCATTTG | GTTAGCTCTTTCAGATTTAC* | 205 |
| Exon 23-2 | GAAGGAAAGAGATACAGAAT* | CCCGCCCGTTCCATAAACTAACAAGCAC | 219 |
| Exon 24 | GTGCTTGTTAGTTTATGGAA* | CGCCCAAAAATTAACTATATTGTGC | 265 |
| Exon 25-1 | CTTGCATCTTAAAATTCATC* | GGTTGCTTGCAGCAATTAAC | 221 |
| Exon 25-2 | CCCGCCCTGGCAATAAAGTTTTGGAT | CAAAATGTGTGGTGATGCTG* | 283 |
| Exon 26 | GGAAATACTTTTGGAAACAT* | TTCCTTGAGTTTACATTAAC | 282 |
| Exon 27-1 | ATGATAGGCTACGTTTTCAT* | TTGCAGTTCTTTTGGTCATC | 220 |
| Exon 27-2 | GTCTTGTAAAGGGGAGAAAG* | TGAGGAGAATTCAGTTCTT | 256 |
| Exon 27-3 | CTGCACAGAAGGCATTTCAG* | CAGAGATGTAGTACAACGTC | 333 |
| Exon 27-4 | ACCAGTTCAGAAGATTATCT | GTCAATAATTTATTGTCGCC* | 256 |
*GC clamp sequence (CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCG) is attached to this site.
Genomic DNA was amplified using 100–200 ng of template DNA, 10 pmol of the mixture of 60 mer primers, 1mM MgCl, and 1 U Platinum Taq in 50 μl PCR reactions. The amplification conditions in a Robocyler (Stratagene, Canada) were five minutes at 94°C (hot start), followed by five cycles of one minute at 94°C, one minute at 52°C, and one minute at 72°C; five cycles of one minute at 94°C, one minute at 50°C, and one minute at 72°C; and 30 cycles of one minute at 94°C, one minute at 48°C, and one minute at 72°C; with five minutes extra at 72°C. The final incubation was followed by five minutes at 94°C and 15 minutes at 50°C to enhance the formation of heteroduplex molecules.
Denaturing gradient gel electrophoresis
Primers for DGGE were obtained from Ingeny (Goes, the Netherlands). Aliquots (4–6 μl) of the PCR products were mixed with 2 μl of standard dye loading buffer and electrophoresed through a 20 cm 9% polyacrylamide gel (acrylamide/bisacrylamide, 37.5/1) containing a linear 20–65% denaturing gradient (100% UF: 7M urea/40% deionised formamide). A 12% polyacrylamide stacking gel was poured to create solid slots for efficient loading of the PCR products, which prevented difficulties caused by the high urea concentration. Electrophoresis was performed in 0.5 Tris acetate EDTA buffer at 58°C for 16 hours.15 Gels were stained with ethidium bromide and photographed under an ultraviolet transilluminator.
To increase the throughput of the DGGE procedure, three to four different amplicons with relatively large melting temperature differences were pooled. The fragment pool was designed using a computer program that was based around melting profiles and sequences (tables 3 and 4). For each different exon, seven samples from four different families were loaded side by side to compare their band patterns. The aberrantly migrating samples were reamplified using sequencing primers and sequencing was performed using the Big Dye cycle sequencing kit according to the manufacturer’s instructions. The reaction products were analysed using an ABI DNA sequencer 310 and sequence files were edited using the Sequence Navigator program.
Table 3.
Amplicon pools created for BRCA1 to increase the throughput of denaturing gradient gel electrophoresis
| Gene | Pool | Exons in amplicon pool |
| BRCA1 | A | 7,18, 10, 20 |
| B | 9, 6, 8, 23 | |
| C | 5, 2B, 11-3, 16B | |
| D | 14, 11-4, 22, 24 | |
| E | 2A, 11-1, 16A | |
| F | 19, 13, 21 | |
| G | 17, 11-14, 3 | |
| H | 11-15, 15A, 15B | |
| I | 11-16, 11-2, 12 | |
| BRCA1 exon 11 | A | 11-5, 11-8, 11-10 |
| B | 11-13b, 11-13e, 11-13f | |
| C | 11-7, 11-12, 11-13c | |
| D | 11-9, 11-11, 11-13a | |
| E | 11-6, 11-13d |
Each amplicon pool contained three to five amplicons with relatively large melting temperature differences.
Table 4.
Amplicon pools created for BRCA2 to increase the throughput of denaturing gradient gel electrophoresis
| Gene | Pool | Exons in amplicon pool |
| BRCA2 | A | 11-1, 27-1, 19 |
| B | 23-1, 18-1, 10-4, 11-3 | |
| C | 8, 20, 27-3, 10-5, 21-1 | |
| D | 15-1, 23-2, 26, 10-6 | |
| E | 12, 10-7, 2, 24, 27-2 | |
| F | 5, 6, 11-17, 15-2 | |
| G | 11-16, 9, 27-4, 18-2 | |
| H | 10-1, 10-3, 10-10, 10-8 | |
| I | 25-1, 14-2, 22-1, 25-2 | |
| J | 14-3, 10-9, 22-2 | |
| K | 4, 3, 7 | |
| L | 16, 11-2, 21-2 | |
| M | 13, 17, 14-1, 10-2 | |
| BRCA2 exon 11 | A | 11-4, 11-8, 11-15h |
| B | 11-5,11-10,11-13, 11-15f | |
| C | 11-6, 11-12, 11-15d | |
| D | 11-7, 11-14, 11-15c | |
| E | 11-9, 11-15g, 11-15m | |
| F | 11-15e, 11-15L, 11-15n | |
| G | 11-15a, 11-15i, 11-15j | |
| H | 11-11, 11-15b, 11-15k |
Each amplicon pool contained three to five amplicons with relatively large melting temperature differences.
RESULTS
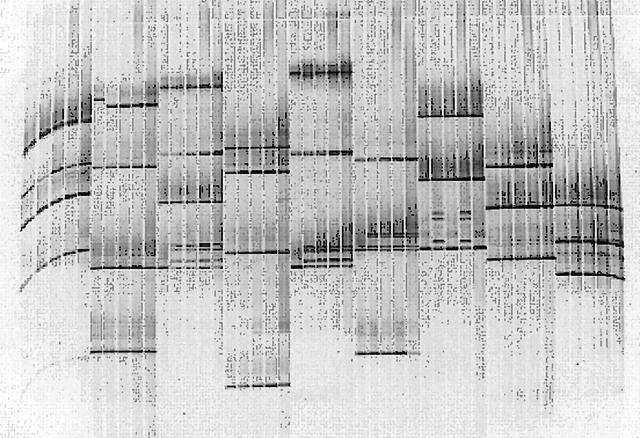
In the 11 patients, aberrantly migrating bands (fig 2) were found in exons 13 and 16 of the BRCA1 gene, and in exons 2, 11, and 14 of the BRCA2 gene (table 5). We then sequenced the exons with aberrantly migrating bands (fig 3). The aberrantly migrating bands found in exons 13 and 16 of BRCA1 and exons 2 and 14 of BRCA2 appeared to be caused by single nucleotide substitutions producing the same amino acid. Consultation with the Breast Cancer Information Core (last accessed 1 February 2005; http://www.research.nhgri.nih.gov/projects/bic) revealed that these substitutions were indeed established polymorphisms.
Figure 2.
Pooling of five samples of 24 exons in the BRCA1 gene for denaturing gradient gel electrophoresis. Each pool consists of three to four amplicons selected on the basis of their melting behaviour. The fragments for each pool can be seen in table 3. The aberrant bands appeared in fragment 9 (pool B), fragment 16B (pool C), fragments 2A and 16A (pool E), fragment 13 (pool F), fragment 3 (pool G), and fragment 11-15 (pool H); these fragments were further analysed by sequencing.
Table 5.
Mutations and polymorphisms found in the BRCA1 and BRCA2 genes using the proposed method
| Patient | Exon, gene | NT | Codon | Base change | AA change | Designation | Mutation type |
| B-III-5 | 11, BRCA2 | 2699 | 824 | Del TAAATG | Leu to stop | 2699 del TAAATG | Nonsense |
| A-III-9 | 13, BRCA1 | 4427 | 1436 | T to C | Ser to Ser | 4427T/C | Polymorphism |
| B-III-5 | |||||||
| D-III-9 | |||||||
| A-III-9 | 16, BRCA1 | 4956 | 1613 | A to G | Ser to Gly | S1613G | Polymorphism |
| B-III-5 | |||||||
| D-III-9 | |||||||
| D-III-9 | 2, BRCA2 | 203 | – | G to A | 5′UTR | 203 >A | Polymorphism |
| D-III-9 | 14, BRCA2 | 7470 | 2414 | A to G | Ser to Ser | 7470A/G | Polymorphism |
AA, amino acid; NT, nucleotide; UTR, untranslated region.
Figure 3.
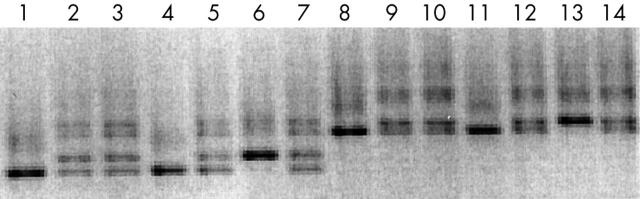
Results of denaturing gradient gel electrophoresis (DGGE) analysis of BRCA1. DGGE of Indonesian patients at high risk for hereditary breast cancer. Exon 16: lanes 1, 4, and 6 displayed a darker single band as opposed to samples in lanes 2, 3, 5, and 7, which showed lighter double bands. The dark band was in the same position as one of the double bands. Exon 13 was similar: lanes 8, 11, and 13 displayed a dark single band, whereas lanes 9, 10, 12, and 14 showed light double bands. These aberrant bands appeared to represent single nucleotide substitutions, with no consequences for the amino acid sequence of the protein (polymorphisms).
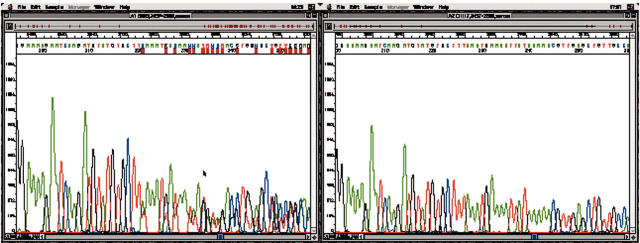
We detected one deleterious BRCA2 mutation in patient B-III-5 (fig 4). This mutation was a deletion of six nucleotides in exon 11 (c.2472delTAAATG) at codon 824. The mutation did not cause a shift of the reading frame, but affected three codons and resulted in a premature stop codon. There is no such mutation registered in the Breast Cancer Information Core (http://www.research.nhgri.nih.gov/projects/bic), so this is a novel mutation. This patient had early onset breast cancer and there was no family history of cancer. Table 5 summarises the genetic aberrations found, and table 6 shows the clinicopathological data of the patients with breast cancer.
Figure 4.
Sequencing traces of BRCA2 exon 11 of a 30 year old patient with early onset breast cancer, showing the 2699delTAAATG mutation on the left, with the normal control on the right.
Table 6.
Clinicopathological data of the patients with breast cancer showing BRAC1/2 mutations and/or polymorphisms
| Patient | Age | Tumour type | Tumour size | TNM | Stage | Lymph nodes positive | Pregnancies | Children | Contraceptives |
| A | 44 | Ductal | 2 | T2N0M0 | II A | 0/11 | 0 | 0 | None |
| B | 30 | Ductal | 1 | T1N0M0 | I | 0/10 | 0 | 0 | None |
| C | 39 | Ductal | 3 | T2N0M0 | II A | 0/10 | 5 | 3 | IUD |
| D | 30 | Ductal | 2.5 | T2N0M0 | II A | 0/10 | 2 | 2 | Oral contraceptives for 4 years, IUD |
IUD, intrauterine device.
DISCUSSION
The increasing demand for mutation detection in disease genes, either known or presumed, can be solved by automated sequencing using fluorescent dyes.16 However, only a few laboratories are equipped for the broad application of this costly and labour intensive strategy. As alternatives to sequencing methods, which determine the exact nature and location of each base along a DNA fragment, various mutation scanning procedures have been developed. These methods, which rely on the recognition of a sequence variation between mutant and wild-type DNA on the basis of an altered electrophoretic migration pattern, provide a simple means for determining whether a given DNA sample harbours a mutation in a particular gene.
The most well established scanning procedures are single strand conformational polymorphism (SCCP) analysis, DGGE, chemical cleavage of mismatch, RNase cleavage, the protein truncation test (PTT), and heteroduplex analysis. Among these methods, SCCP, DGGE, PTT, and heteroduplex analysis are the most widely used because of their accuracy, simplicity, lack of toxicity, and/or relative affordability.
We focused on DGGE because this method, when optimised, has the highest mutation detection rate (close to 100%17) compared with SCCP and heteroduplex analysis. Additional advantages of this methodology are the possibility of optimising the analysis by computer simulation and the non-radioactive approach. In DGGE, during electrophoresis, double stranded DNA amplified by PCR migrates through a gel containing an increasing concentration of denaturant (urea and formamide). As double stranded DNA is electrophoresed through the denaturing gradient, it will melt and change its conformation in such a way that the mobility of the molecule is dramatically reduced. To prevent complete strand dissociation and to facilitate the detection of mutations in the higher melting domains, a GC rich fragment (GC clamp) is introduced during fragment amplification. The GC clamp increases the percentage of single base changes detectable by DGGE, theoretically to 100%.18,19 In principle, four bands are detectable in a heterozygous state after denaturation and renaturation, corresponding to the two homodimers and two heterodimers. Although DGGE has been applied to BRCA mutation screening before,20–22 we used a new technique based on DGGE after exon by exon PCR amplification of the complete BRCA1 and BRCA2 sequences,12 and sequenced aberrantly moving bands. This yielded four polymorphisms consisting of single nucleotide substitutions, underlining the sensitivity of the method. Intelligent pooling of the amplicons before electrophoresis greatly improved the throughput of the method.
Double gradient DGGE is based on the combination of two linear gradients, a primary denaturing gradient (urea and formamide) and a collinear secondary porosity gradient (polyacrylamide).23 This secondary gradient suppresses band broadening during electrophoresis and thus improves the resolution of the DGGE banding pattern. Hayes et al compared double gradient DGGE gels with various porosity gradients to a standard 9% polyacrylamide gel, and showed that mutations with different melting profiles cannot be appropriately detected using a single DGGE condition.15
PTT is a widely applied screening technique,5 but is especially helpful for known mutations in particular populations, and less suitable as a primary screening approach for new mutations in unknown populations. Furthermore, PTT only detects mutations that result in stop codons and lead to premature termination of translation, thereby producing truncated proteins. A possible advantage of PTT is that it conveniently misses harmless polymorphisms. PTT is usually only applied for detecting mutations in exon 11 of the BRCA1 gene and exons 10 and 11 of the BRCA2 gene, which account for more than 60% of the coding sequence. However, mutations are distributed throughout the entire coding sequence, with no apparent clustering or hot spots.24
“Intelligent pooling of the amplicons before electrophoresis greatly improved the throughput of the method”
More than 1893 distinct germline BRCA2 mutations have been found to date (http://www.research.nhgri.nih.gov/projects/bic), and the number is expected to increase further. The pattern of mutations is similar to that seen in BRCA1 and in many other tumour suppressor genes. At present, all mutations that clearly cause disease result in premature termination of translation or absence of a transcript. Approximately 75% of truncating mutations are small deletions, 15% are small insertions, and 10% are base substitutions leading directly to termination codons (http://www.research.nhgri.nih.gov/projects/bic).25–27 In addition, the novel 2699delTAAATG mutation described in our present paper leads to a premature stop codon. The patient with this mutation had early onset breast cancer and no family history of cancer. It has been suggested that the probability of harbouring a BRCA2 mutation among patients with early onset breast cancer is relatively independent of a positive family history for the disease.25
Take home messages.
Four BRCA1/2 polymorphisms and one novel BRCA2 mutation were found in a group of Indonesian patients at high risk of hereditary breast cancer using pooled denaturing gradient gel electrophoresis and targeted sequencing
This new and high throughput method is sensitive and particularly suited for screening unknown populations
Mutations in the BRCA2 gene have been found mainly in families with a high incidence of female and male breast cancer,4,26,28,29 whereas the risk of ovarian cancer is lower than in BRCA1 families.4,26,30,31 Loss of heterozygosity has been found in 30–45% of sporadic breast tumours,32,33 indicating a role for BRCA2 also in sporadic breast tumours, although very few somatic mutations have been reported.24,34,35 Other tumour types seen repeatedly in BRCA2 families include those of the prostate, larynx, pancreas, and colon.36,37
In conclusion, we describe a novel BRCA2 mutation and four polymorphisms in the BRCA1/2 genes found in the Indonesian population using a new high throughput and highly sensitive method to screen BRCA1 and BRCA2 for mutations based on exon by exon PCR amplification followed by pooled DGGE and sequencing of aberrant bands. This is an ideal approach for screening populations for unknown mutations, and has the ability to detect single base differences using non-toxic and relatively simple and inexpensive methods.
Acknowledgments
Supported by grant IN-2001-008 of the Dutch Cancer Society. We thank Dr J Hilgers who was instrumental in setting up the Familial Cancer Clinic initiative in Jogjakarta.
Abbreviations
DGGE, denaturing gradient gel electrophoresis
PCR, polymerase chain reaction
PTT, protein truncation test
SCCP, single strand conformational polymorphism
The patients gave full consent for their details to be published.
REFERENCES
- 1.Jardines L, Haffty BG, Doroshow JH, et al. Breast cancer overview. In: Cancer management: a multidisciplinary approach. 2001:137–59.
- 2.Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 1990;250:1684–9. [DOI] [PubMed] [Google Scholar]
- 3.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994;266:66–71. [DOI] [PubMed] [Google Scholar]
- 4.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility BRCA2. Nature 1995;378:789–92. [DOI] [PubMed] [Google Scholar]
- 5.Zweemer RP, Verheijen RHM, Gille JJP, et al. Clinical and genetic evaluation of 30 ovarian cancer families. Am J Obstet Gynecol 1998;178:85–90. [DOI] [PubMed] [Google Scholar]
- 6.Zweemer RP, van Diest PJ, Verheijen RH, et al. Molecular evidence linking primary cancer of the fallopian tube to BRCA1 germline mutations. Gynecol Oncol 2000;76:45–50. [DOI] [PubMed] [Google Scholar]
- 7.Narod SA, Ford D, Devilee P. An evolution of genetic heterogeneity in 145 breast-ovarian cancer families. The breast cancer linkage consortium. Am J Hum Genet 1995;56:254–64. [PMC free article] [PubMed] [Google Scholar]
- 8.Breast Cancer Linkage Consortium. Pathology of familial breast cancer: differences between breast cancer in carriers of BRCA1 or BRCA2 mutations and sporadic cases. Lancet 1997;449:1505–10. [PubMed] [Google Scholar]
- 9.Liede A, Narod SA. Hereditary breast and ovarian cancer in Asia: genetic epidemiology of BRCA1 and BRCA2. Hum Mutat 2002;20:413–24. [DOI] [PubMed] [Google Scholar]
- 10.Neuhausen SL. Ethnic differences in cancer risk resulting from genetic variation. Cancer 1999;86:2575–82. [DOI] [PubMed] [Google Scholar]
- 11.Verhoog LC, van den Ouweland AM, Berns E, et al. Large regional differences in the frequency of distinct BRCA1/BRCA2 mutations in 517 Dutch breast and/or ovarian cancer families. Eur J Cancer 2001;37:2082–90. [DOI] [PubMed] [Google Scholar]
- 12.Van der Hout AH, Mulder IM, Wu Y, et al. A DGGE system for comprehensive mutation screening of the complete coding regions of BRCA1 and BRCA2 outside exons 11. Am J Hum Genet 1999;65 (suppl S Oct) :2322. [Google Scholar]
- 13.Fischer SG, Lerman LS. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A 1983;80:1579–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moller P, Evans G, Haites N, et al. Guidelines for follow-up of women at high risk for inherited breast cancer: consensus statement from the Biomed 2 demonstration programme on inherited breast cancer. Dis Markers 1999;15:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes VM, Wu Y, Osinga J, et al. Improvements in gel composition and electrophoretic conditions for broad-range mutation analysis in denaturing gradient gel electrophoresis. Nucleic Acid Res 1999;27:e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenthal A, Chamock JDS. New protocols for DNA sequencing with dye terminators. DNA Seq 1992;3:61–4. [DOI] [PubMed] [Google Scholar]
- 17.Macek M Jr, Mercier B, Mackova A, et al. Sensitivity of the denaturing gel electrophoresis technique in detection of known mutations and a novel Asian mutation in the CFTR gene. Hum Mutat 1997;9:136–47. [DOI] [PubMed] [Google Scholar]
- 18.Abrams ES, Murdaugh SE, Lerman LS. Comprehensive detection of single base changes in human genomic DNA using denaturing gradient gel electrophoresis and a GC-clamp. Genomics 1990;7:463–75. [DOI] [PubMed] [Google Scholar]
- 19.Sheffield VC, Cox DR, Lerman LS, et al. Attachment of a 40-base pair G-C rich sequence (GC clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A 1989;86:232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller D, Bonatiti-Pelie C, Abecassis J, et al. BRCA1 testing in breast and/or ovarian cancer families from northeastern France identifies two common mutations with a founder effect. Fam Cancer 2004;3:15–20. [DOI] [PubMed] [Google Scholar]
- 21.Gal I, Gershoni Baruch R, Haber D, et al. The 1100delAT BRCA1 and the 8765delAG BRCA2 mutations: occurrence in high-risk non-Ashkenazi Jews and haplotype comparison of Jewish and non-Jewish carriers. Fam Cancer 2004;3:11–14. [DOI] [PubMed] [Google Scholar]
- 22.Atoum MF, Al-Kayed SA. Mutation analysis of the breast cancer gene BRCA1 among breast cancer Jordanian females. Saudi Med J 2004;25:60–3. [PubMed] [Google Scholar]
- 23.Gelfi C, Righetti SC, Zunino F, et al. Detection of p53 mutations by double-gradient gel electrophoresis. Electrophoresis 1997;18:2921–7. [DOI] [PubMed] [Google Scholar]
- 24.Shattuck-Eidens D, Mcclure M, Simard J, et al. A collaborative survey of 80 mutations in the BRCA1 breast and ovarian cancer susceptibility gene. JAMA 1995;273:535–41. [PubMed] [Google Scholar]
- 25.Plaschke J, Commer T, Jacobi C, et al. BRCA2 germline mutations among early onset breast cancer patients unselected for family history of the disease. J Med Genet 2000;37:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couch FJ, Farid LM, DeShano ML, et al. BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet 1996;13:123–5. [DOI] [PubMed] [Google Scholar]
- 27.Teng DH-F, Bogden R, Mitchell J, et al. Low incidence of BRCA2 mutations in breast carcinoma and other cancer. Nat Genet 1996;13:241–4. [DOI] [PubMed] [Google Scholar]
- 28.Phelan CM, Lancaster JM, Tonin P, et al. Mutation analysis of the BRCA2 gene in 49 site-specific breast cancer families. Nat Genet 1996;13:120–2. [DOI] [PubMed] [Google Scholar]
- 29.Tavtigian SV, Simard J, Rommens J, et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet 1996;12:333–7. [DOI] [PubMed] [Google Scholar]
- 30.The Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a large population based series of breast cancer cases. Br J Cancer 2000;83:1301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antoniou AC, Gayter SA, Stratton JF, et al. Risk models for familial ovarian and breast cancer. Genet Epidemiol 2000;18:173–90. [DOI] [PubMed] [Google Scholar]
- 32.Clenton-Jansen A-M, Collins N, Lakhani SR, et al. Loss of heterozygosity in sporadic breast tumours at the BRCA2 locus on chromosome 13q12–q13. Br J Cancer 1995;72:1241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van den Berg J, Johannsson O, Hakansson S, et al. Allelic loss at chromosome 13q12–q13 is associated with poor prognosis in familial and sporadic breast cancer. Br J Cancer 1996;74:1615–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lancaster JM, Wooster R, Mangion J, et al. BRCA2 mutations in primary breast and ovarian cancer. Nat Genet 1996;13:238–40. [DOI] [PubMed] [Google Scholar]
- 35.Miki Y, Katagiri T, Kasumi F, et al. Mutation analysis in the BRCA2 gene in primary breast cancer. Nat Genet 1996;13:245–7. [DOI] [PubMed] [Google Scholar]
- 36.The Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst 1999;91:1310–16. [DOI] [PubMed] [Google Scholar]
- 37.Basham VM, Lipscombe JM, Ward JM, et al. BRCA1 and BRCA2 mutations in a population-based study of male breast cancer. Br Cancer Res 2002;4:R2 Epub 2001 Nov 21. [DOI] [PMC free article] [PubMed] [Google Scholar]