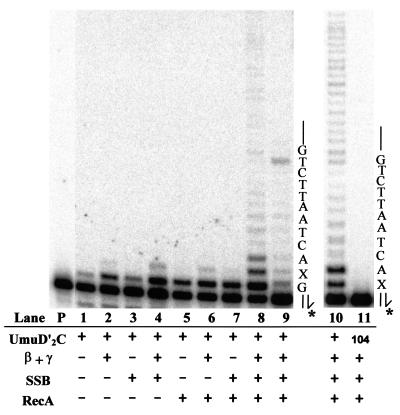
Figure 3.
Protein cofactor requirements for UmuD′2C (E. coli pol V)-catalyzed lesion bypass. Each reaction in lanes 1–9 contains 1.5 μl of Superdex 200 fx 64 containing UmuD′2C (having no detectable pol IIIts), pol I antibody, and various combinations of RecA, β, γ complex, and SSB, as indicated (lanes 1–9). Running-start reactions, in which C is incorporated opposite template G to reach the abasic site, were carried out at 37°C, with all four dNTPs present in lanes 1 to 8, but with dCTP omitted in lane 9. Reactions were run in the presence of 5% polyethylene glycol. A portion of the template sequence is shown at the right of lane 9, where X represents an abasic site. The left-hand lane contains the primer (P) in the absence of proteins. Standing-start reactions, in which the first incorporated nucleotide occurs opposite X, were run for wild-type UmuD′2C (lane 10) and for the mutant UmuD′2C104 (D101N) (lane 11), each at a concentration of 200 nM. A portion of the template, shown at the right of lane 11, has the same sequence as the running-start template, but uses a primer terminating one base before the lesion. The asterisk (*) designates a 32P-label at the 5′-primer terminus. The running-start primer-template DNA is shown at the top of Fig. 2.