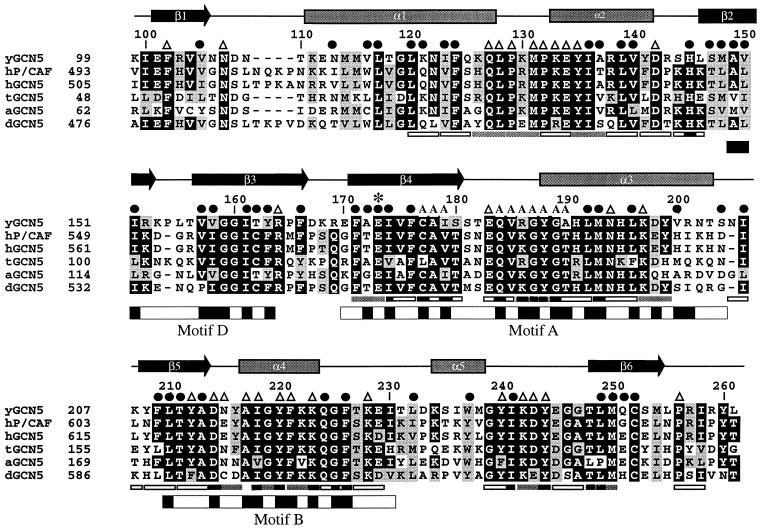
Figure 2.
Sequence alignment of the GCN5 family of HAT domains. The primary sequence of the yeast GCN5 (yGCN5) HAT domain used for the structure determination is shown at the top of the alignment. Sequences from the homologous HAT domains from GCN5 of Arabidopsis, Drosophila, human, and Tetrahymena as well as with human P/CAF are aligned [clustal program (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalW/)] and displayed [boxshade program (http://www.ch.embnet.org/software/BOX_form.html)]. Black and gray backgrounds are used to indicate identical and/or conserved residues found in at least 50% of the proteins at a given position, respectively. Secondary structural elements within the HAT domain of yGCN5 are shown above the sequence alignment. ●, residues that are buried within the core of the protein; A, residues that are implicated from the HAT1 and SmAAT structures to contact the CoA cofactor (53); ▵, solvent exposed residues that are presumably accessible for coenzyme or substrate binding and catalysis; ∗, the position of the putative general base (Glu173) for histone acetylation. Positions of alanine mutations that decrease HAT activity are indicated below the sequence alignment: Triple mutations are indicated with gray bars (38), and single mutations are indicated with black bars (37). Triple alanine mutations that have negligible effect on HAT activity are indicated with open rectangles (all of the single mutations are contained within the triple mutations). The GNAT conserved motifs D, A, and B (39) are indicated below the sequence alignment, with black shading indicating regions of high sequence homology within the GNAT superfamily.