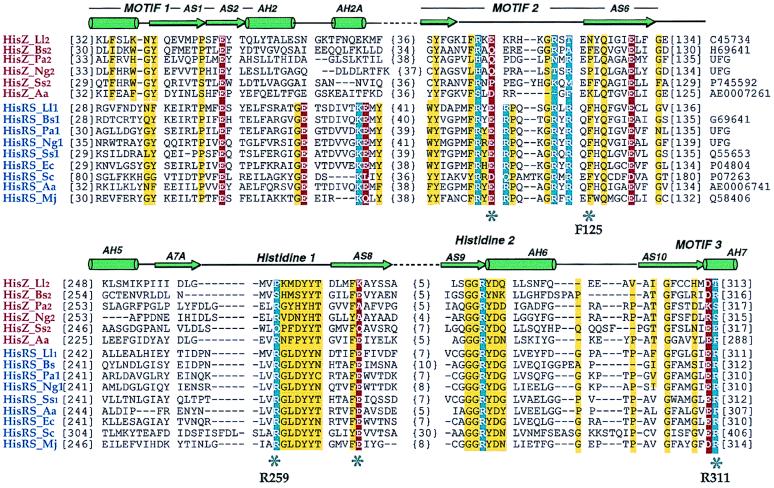
Figure 1.
Structure-based sequence alignment of HisRS-like proteins (HisZ) and representative functional histidyl-tRNA synthetases (HisRSs). Species names are abbreviated as follows: Ll, L. lactis; Bs, B. subtilis; Pa, P. aeriginosa; Ng, N. gonorrhoeae; Ss, Synechocystis sp.; Aa, A. aeolicus; Ec, E. coli; Sc, Saccharomyces cerevisae; Mj, Methanococcus jannaschii. Numbers at the left of the sequences indicate positions of first aligned residues in the block, whereas the bracketed numbers indicate spacing between the elements of the blocks. Swiss-Prot, Protein Identification Resource, and National Center for Biotechnology Information (NCBI)/GenBank accession numbers are indicated to the right. UFG indicates sequences retrieved from the unfinished genomes of the NCBI server. The three diagnostic class II motifs and HisRS-specific peptide motifs are indicated and are overlaid by a schematic representation of HisRS secondary structure (25, 28). Highly conserved neutral, acidic, and basic residues are outlined in yellow, red, and blue, respectively; and key residues (E. coli HisRS numbering) that denote significant differences between the HisRS functional and HisRS-like proteins are labeled.