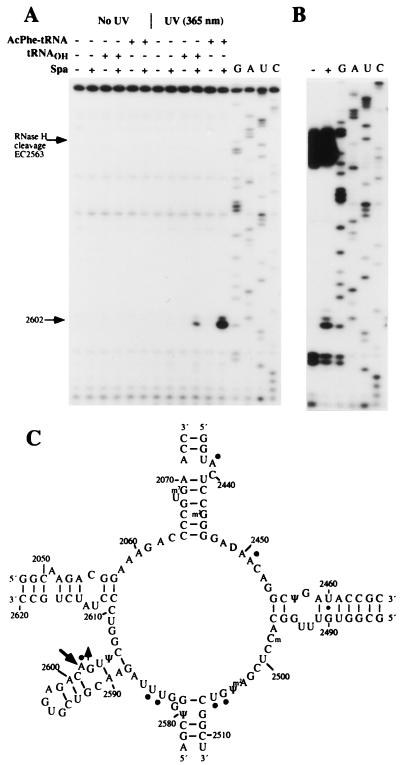
Figure 2.
Primer extension analysis of sparsomycin-rRNA crosslinks. (A) The autoradiograph shows primer extension from EC2621, and the stop at A2602 is indicated. Control samples with no irradiation, and no sparsomycin, are included. 70S ribosomes (0.15 μM) and poly(U) (1 μg/pmol ribosome) were mixed with sparsomycin in the presence, or absence, of P/P′-site-bound tRNA (0.18 μM) (see Materials and Methods) and were irradiated at 365 nm for 15 min. G, A, U, and C represent sequencing tracks. (B) 70S ribosomes.poly(U).N-Ac-Phe-tRNA complexes were irradiated at 365 nm as described above, and the extracted RNA was cleaved by RNase H in the presence of oligonucleotides EC2563 and EC2654. The resulting ≈83-nt fragment was gel-purified and analyzed by primer extension from EC2621. The band at A2602 is indicated. (C) Secondary structure of the peptidyl transferase loop region of 23S rRNA from E. coli showing the crosslinked site (large arrow) and the nucleotides that display reduced (circles), or enhanced (circle plus arrow), chemical reactivities with N-Ac-Phe-tRNA in the P/P′-site (8).