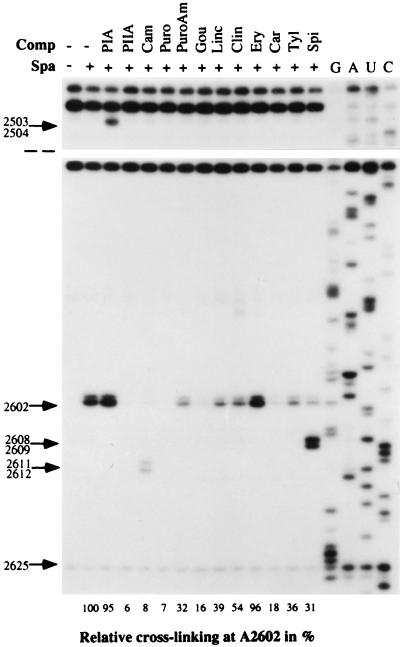
Figure 7.
Interference of peptidyl transferase antibiotics with the sparsomycin crosslink at A2602. Sparsomycin (50 μM) was complexed to E. coli 70S ribosome.poly(U) complexes in the presence of N-Ac-Phe-tRNA and was incubated at 37°C for 5 min. The second drug was added, and, after 15 min of incubation, the samples were irradiated for 15 min at 365 nm. Primer extension analyses were performed by using EC2654. Drug concentrations were pristinamycin IA (PIA, 200 μM), pristinamycin IIA (PIIA, 200 μM), chloramphenicol (Cam, 1.2 mM), puromycin (Puro, 1 mM), puromycin aminonucleoside (PuroAm, 1 mM), gougerotin (Gou, 200 μM), lincomycin (Linc, 200 μM), clindamycin (Clin, 200 μM), erythromycin (Ery, 200 μM), carbomycin (Carb, 200 μM), tylosin (Tyl, 200 μM), and spiramycin (Spi, 200 μM). The primer extension stop at A2602 was quantified in an Instant Imager, using the stop at 2625 as a reference, and the results are given as a percentage below the gel, relative to a control sample in the absence of competitor. Control experiments, performed in the absence of sparsomycin, established that the observed effects depended on the presence of sparsomycin, except for pristinamycin IA and chloramphenicol (see text). G, A, U, and C denote sequencing tracks.