Abstract
Background and aims: Genetic variation in NOD2 has been associated with susceptibility to Crohn’s disease (CD) and specifically with ileal involvement. The reason for the unique association of NOD2 mutations with ileal disease is unclear. To identify a possible link, we tested expression of NOD2 in intestinal tissue of CD patients and controls.
Patients and methods: Fifty five specimens of ileum or colon from 21 CD patients, seven ulcerative colitis (UC) patients, and five controls with pathology other than CD or UC were stained for NOD2 using an immunoperoxidase method.
Results: Using a monoclonal antibody against NOD2 developed in our laboratory, we detected uniform expression of NOD2 in terminal ileum Paneth cells from controls and patients as well as in metaplastic Paneth cells in the colon. Mechanical purification showed enriched expression of NOD2 mRNA in ileal crypts. In Paneth cells, NOD2 was located in the cytosol in close proximity to the granules that contain antimicrobial peptides. We detected minimal NOD2 in the villous epithelium of the ileum or in the colonic epithelium from both CD patients and controls.
Conclusions: These results suggest a role for NOD2 in the regulation of Paneth cell mediated responses against intestinal bacteria and a plausible mechanism to explain the selective association of NOD2 mutations with ileal disease. The impaired capacity of CD associated mutations to sense luminal bacteria may result in increased susceptibility to certain gut microbes.
Keywords: NOD2, Crohn’s disease, Paneth cell, innate immunity
Crohn’s disease (CD) is a chronic relapsing inflammatory disease of the gastrointestinal tract most commonly affecting the terminal ileum and colon with a prevalence of approximately 100–200/100 000 in Western countries.1,2 CD is considered a multifactorial disease in which both genetic and environmental factors appear to play a role in disease development.3 A role for intraluminal bacteria in disease pathogenesis is suggested by the absence of inflammation in various genetic models of colitis when animals are raised in germ free condition.3 Furthermore, antibiotic therapies can be effective in the treatment of CD.4
Recent studies have revealed coding region polymorphisms in NOD2 which are significantly associated with susceptibility to CD.5–7 The NOD2 product is a protein composed of two NH2 terminal caspase recruitment domains (CARDs), a centrally located nucleotide binding domain, and multiple COOH terminal leucine rich repeats.8 Three major polymorphisms in NOD2, L1007fsinsC, G908R, and R702W, have been genetically associated with CD in European and American populations.5–7 Genotype analyses of patients have shown that the NOD2 variants are more significantly associated with ileal disease but much less or not at all with colon predominant disease.9–11 Functional studies have shown that NOD2 activates nuclear factor κB (NFκB) via the serine-threonine kinase RICK8 and confers responsiveness to bacterial lipopolysaccharide (LPS) and peptidoglycan (PGN).12 NOD2 variants associated with CD are deficient in their ability to activate NFκB in response to LPS and PGN.8,13 Recent studies have revealed that muramyl dipeptide derived from PGN is the structure in bacteria recognised by NOD2,14,15 and induction of NFκB activation is deficient in mononuclear cells derived from CD patients homozygous for L1007fsinsC.14 NOD2 has been shown to be expressed in myelomonocytic cells, further suggesting a role for this protein in innate immunity against bacteria.8,16 Although the precise function of NOD2 is not entirely understood, these studies suggest that NOD2 acts in a defence response against bacteria and a deficit in NOD2 activity is associated with disease. Notably, NOD2 mutations are primarily associated with ileal disease.9–11 However, the underlying mechanism to explain the link betweeb NOD2 mutations and ileal involvement is not understood.
Paneth cells are specialised intestinal cells located primarily at the base of the crypts of Liberkühn in the small intestine. They secrete defensins and other antimicrobial peptides in response to bacteria.17–19 Paneth cells are part of the host defence against luminal bacteria and are thought to release their secretory granules into the narrow lumen of the crypt to protect the epithelial stem cells from microbes. Paneth cells have been suggested to play a role in CD17 but clear evidence for a direct role in disease pathogenesis is lacking. Recent studies however have shown lower expression of human β-defensin 1 in intestinal tissue in CD compared with ulcerative colitis (UC).20 However, additional studies are needed to establish a role for defensins in CD. The mechanism by which Paneth cells regulate host defences are poorly understood but they are likely to involve, at least in part, secretion of antimicrobial products in response to bacterial products, including muramyl dipeptide, the molecule recognised by NOD2.19 Here we report that NOD2 is expressed in Paneth cells, a finding that suggests a role for these cells in the selective association of NOD2 mutations with ileal disease.
MATERIALS AND METHODS
Expression plasmids, immunoblotting, and immunoprecipitation
Expression plasmids for NOD1, NOD2, Apaf-1, and RICK have been described.8,12 Ipaf and Pypaf-1 cDNAs were generated by polymerase chain reaction (PCR) and cloned into the pcDNA-Flag plasmid. Expression plasmids producing N terminal HA tagged NOD2 variants P268S, P268S/R702W, and P268S/G908R were generated by the QuikChange XL site directed mutagenesis kit (Stratagene, La Jolla, California, USA). P268S/L1007fsinsC were generated by a PCR method using P268S DNA as templates. The generated PCR products were cloned into the pMX-puro expression plasmid. The authenticity of all constructs was confirmed by DNA sequencing.13 Transfection of expression constructs into HEK293T cells was performed using the calcium phosphate method, as described previously.8 Expression from expression plasmids in transfected cells was determined by immunoblotting using monoclonal anti-HA antibody, 16B12 (Covance, La Jolla, California, USA), anti-Flag antibody, M2 (Sigma, St Louis, Missouri, USA), anti-Myc antibody, 9E10 (Santa Cruz Biotechnology, Santa Cruz, California, USA), as described previously.8 For immunoprecipitation assay, proteins immunoprecipitated with anti-NOD2 or control monoclonal antibody, anti-Flag antibody, M2, were detected by immunoblotting with rabbit polyclonal anti-NOD2 antibody, as described previously.5,8
Monoclonal anti-NOD2 antibody generation
Recombinant human NOD2 protein (residues 28–301) was overexpressed in Escherichia coli strain BL21 Star (DE3) pLysS (Invitrogen, Carlsbad, California, USA) using the pET-30a vector (Novagen, Madison, Wisconsin, USA). Recombinant NOD2 protein containing a C terminal histidine tag was purified using a nickel column, His-Bind Resin (Novagen). Six week old BALB/c mice were immunised by intraperitoneal injection of purified NOD2 (10–50 μg/animal) in complete Freund’s adjuvant. Animals were boosted (intraperitoneally) at intervals of 14–28 days with NOD2 protein in incomplete Freund’s until a sufficient titre of antibody was present in venous blood. Four days following the final boost with antigen, splenocytes from an immunised animal were fused with the P3X63-Ag8.653 murine myeloma cells (ATCC) using the method of Kearney and colleagues.21 Hybridoma supernatants were screened for anti-NOD2 activity by enzyme linked immunosorbent assay 14–18 days post fusion. One hybridoma, termed 2D9, remained positive after repeated testing and was selected for further study. 2D9 was purified from ascitis of pristane primed BALB/c mice injected intraperitoneally with 2D9 hybridoma cells using a DEAE sepharose ion exchange column.22
Tissue specimens
Thirty two specimens of ileum or colon from 21 patients with CD (nine males and six females, aged 15–66 years (mean 32)) and 12 specimens of colon from seven patients with UC (two males and five females, aged 16–66 years (mean 42)) from the University of Michigan Hospitals were available for the study. In addition, two CD patients homozygous for the L1007fsinsC NOD2 mutation from the University of Chicago Hospitals were studied. In all cases, the clinical diagnosis was confirmed by pathological studies. Five control specimens of resected ileum or colon for pathologies other than CD or UC from five patients (bowel obstruction or tumour; one male and four females, aged 14–74 years (mean 38)) were also included in the study. Analysis of human tissues was approved by the human research review boards of the University of Michigan and University of Chicago Medical Schools and the Royal Free Hamstead NHS Trust.
Immunohistochemical analyses
Sections (5 µm) of formalin fixed paraffin embedded tissues were mounted on Probe-On slides (Fisher Scientific, Itasca, Illinois, USA), deparaffinised in xylene, and rehydrated in distilled H2O through graded alcohols. In most studies, antigen retrieval was enhanced by microwaving the slides for 10 minutes in 10 mM citrate buffer, pH 6.0 (Biogenex, San Ramon, California, USA). Endogenous peroxidase activity was quenched by incubation with 6% hydrogen peroxide in methanol, and then the sections were washed and blocked with 1.5% normal mouse serum for one hour. Sections were incubated with purified 2D9, a mouse monoclonal anti-NOD2 antibody developed in the present study at 2 μg/ml for two hours at room temperature. Monoclonal antihuman CD68 antibody (Clone PG-M1; Dako Cytomation, Ely, Cambridgeshire, UK) was diluted 1:250 in 1% horse serum in phosphate buffered saline (PBS). Incubation with goat antimouse EnVision horseradish-peroxidase conjugate (Dako, Carpinteria, California, USA) for 30 minutes at room temperature was used as the detection system for antibody binding. All experiments included sections stained with purified isotype matched mouse monoclonal antibody, anti-Schistosoma japonicum glutathione S-transferase antibody, B-14 (Santa Cruz Biotechnology), under the same conditions as anti-NOD2 antibody, to monitor non-specific staining. Immunostained sections were lightly counterstained with methylene blue and then examined by light microscopy.
Isolation of intestinal villus and crypt epithelial cells
The mucosa was dissected from surgically resected segments of terminal ileum, rinsed in 10 mM dithiothreitol in PBS for 10 minutes, and incubated in 30 mM EDTA for 10 minutes at room temperature to detach the epithelium from the basement membrane. Mucosal segments were pinned onto a corkboard, attached to a mechanical shaker, and shaken in aliquots of ice cold PBS for one minute periods. Shaking of terminal ileum mucosal segments initially detached epithelial cells from the villi, and subsequently from crypts, with intervening fractions containing mixed populations, and the composition of each fraction was evaluated microscopically. Relative expression of lysozyme mRNA, determined by quantitative real time reverse transcription (RT)-PCR, was subsequently used to confirm that the villus derived fractions did not contain Paneth cells while crypt derived fractions were greatly enriched for this marker. Epithelial cells were collected by centrifugation at 400 g for five minutes, and lysed in Trizol reagent (Life Technologies Ltd, Paisley, UK) to extract cellular RNA.
Quantitative real time RT-PCR
Total cellular RNA was isolated using Trizol reagent (Life Technologies Ltd) according to the manufacturer’s protocol, and quantified by agarose gel electrophoresis and spectrophotometry. For cDNA synthesis, 2 μg of total RNA and 0.5 μg oligo(dT) primer (Life Technologies Ltd) in 20 μl of nuclease free water were heated to 65°C for five minutes and rapidly cooled on ice. Reverse transcription was performed at 37.0°C for two hours in a 50 μl volume comprising 50 mM Tris HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, 0.5 mM each of dATP, dTTP, dCTP, and dGTP, and 800 U Moloney murine leukaemia virus reverse transcriptase (Life Technologies Ltd).
Quantitative real time PCR was performed using the Rotor-Gene amplification system (Biogene, Kimbolton, UK). Each 20 μl PCR reaction contained 2 μl of cDNA, 1 μM of sense and anti-sense primer, 1.5 mM MgCl2, 5 mM each of dATP, dTTP, dCTP, and dGTP, 10 U SYBR green (Biogene), 2 μl PCR buffer (Qiagen, Crawley, UK), 4 μl of Q-solution (Qiagen), and 0.25 U Taq polymerase (Qiagen). Intron spanning primer pairs were used to amplify each cDNA. The sequences were NOD2: 5′- GCGCGATAACAATATCTCAGA -3′ and 5′- CAGAGTTCTTCTAGCATGACG-3′; lysozyme: 5′-GCTGCAAGATAACATCGCTG-3′ and 5′-GCCATATTTTGCTCCTGCTTC-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5′-CGAGATCCCTCCAAAATCAAG-3′ and 5′-GAGCTTGACAAAGTGGTCG-3′. Relative levels of GAPDH, NOD2, and lysozyme PCR products were calculated according to the threshold cycle (CT) value, normalised using the value of the sample with the lowest level of each product, and the data expressed as the ratio of NOD2 or lysozyme to GAPDH. Specificity of the desired PCR products was determined by melting curve analysis.
RESULTS
Development of a monoclonal antibody specific for human NOD2
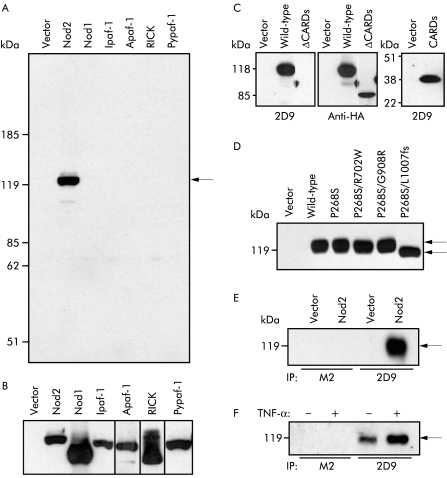
A monoclonal antibody, termed 2D9, was developed against recombinant NOD2 protein (amino acid residues 28–301). To verify the reactivity and specificity of 2D9, we transfected HEK293T cells with constructs expressing NOD2, several NOD2 related proteins including NOD1, Ipaf, Apaf-1, Pypaf-1, RICK, or empty vector, and the cell lysates were analysed with 2D9. Immunoblotting analysis revealed that 2D9 recognised NOD2 but not any of the control proteins tested (fig 1A ▶). Control proteins, including NOD1, Ipaf, Apaf-1, Pypaf-1, and RICK, were appropriately expressed in cell lysates (fig 1B ▶). Analysis of NOD2 mutants lacking CARDs or expressing CARDs alone showed that 2D9 recognises CARDs of NOD2, which is consistent with the fact that amino acid residues 28–301 of NOD2 were used to generate 2D9. All three CD variants, R702W, G908R, and L1007fsinsC, occur on the same haplotype background, which includes the common P268S variant.5,6 To assess whether 2D9 recognises the NOD2 variants associated with CD, we constructed plasmids to express G908R, R702W, and L1007fsinsC proteins in the presence of P268S. Lysates prepared from HEK293T cells transfected with NOD2 constructs were immunoblotted with 2D9. The analysis revealed that 2D9 recognises all NOD2 variants associated with CD (fig 1D ▶). To assess if 2D9 can immunoprecipitate NOD2, extracts from cells transfected with the NOD2 expression plasmid or control plasmid were immunoprecipitated with 2D9 and the immunoprecipitates were immunoblotted with the rabbit anti-NOD2 antibody.5 We detected NOD2 when the cell extracts were immunoprecipitated with 2D9 but not with class matched control antibody, anti-Flag antibody, M2 (fig 1E ▶). Furthermore, endogenous NOD2 protein was specifically immunoprecipitated from the leucocyte cell line HL-60 (fig 1F ▶). Stimulation of HL-60 with tumour necrosis factor α (TNF-α) induced NOD2 protein (fig 1F ▶), which is consistent with the recent observation that NOD2 mRNA levels are upregulated by TNF-α in HL-60 cells.16 These results indicate that 2D9 recognises specifically human NOD2.
Figure 1.
Characterisation of 2D9, a monoclonal anti-NOD2 antibody. (A, B) HEK293T cells were transfected with empty pcDNA3 vector or pcDNA3 vector to express HA tagged Nod2 (114 kD), HA tagged Nod1 (110 kD), HA tagged Ipaf (118 kD), Myc tagged Apaf-1 (138 kD), Flag tagged RICK (63 kD), or Flag tagged Pypaf-1 (120 kD). Cell lysates were immunoblotted with 2D9, a mouse monoclonal anti-NOD2 antibody (A), mouse monoclonal anti-HA antibody (16B12), mouse monoclonal anti-Flag antibody (M2), or mouse monoclonal anti-Myc antibody (9E10) (B). (C) HEK293T cells were transfected with empty pcDNA3 vector or pcDNA3 vector to express HA tagged wild-type Nod2 or mutant Nod2 lacking caspase recruitment domains (ΔCARDs) or expressing CARDs alone (CARDs). Immunoblottings with 2D9 or anti-HA antibody are shown. (D) HEK293T cells were transfected with empty pcDNA3 vector or pcDNA3 vector to express wild-type Nod2 or mutant Nod2, P268S, P268S/R702W, P268S/G908R, or P268S/L1007fs. Cell lysates were blotted with 2D9. (E) HEK293T cells were transfected with empty pcDNA3 vector or pcDNA3 vector to express wild-type Nod2. Cell lysates were immunoprecipitated with 2D9 or M2, an isotype matched control antibody. Immunorecipitates were blotted with rabbit polyclonal anti-Nod2 antibody. (F) HL60 cells were cultured with or without tumour necrosis factor α (TNF-α) for 24 hours. Cell lysates from 2×108 cells were immunoprecipitated with 2D9 or M2. Immunoprecipitates were blotted with rabbit polyclonal anti-Nod2 antibody.
NOD2 is expressed in Paneth cells of the ileum
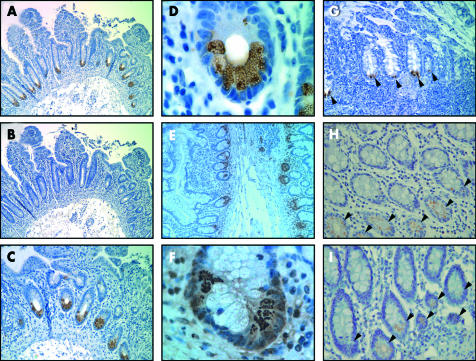
The monoclonal antibody against NOD2 was employed in immunoperoxidase staining of formalin fixed paraffin embedded terminal ileum and colon specimens from CD patients and controls. In the terminal ileum of CD patients and controls, NOD2 was detected in Paneth cells that were identified by their location in crypts and by their prominent granules in cytoplasm (fig 2A ▶, B). Expression of NOD2 in Paneth cells was found in both uninvolved (fig 2A ▶) and inflamed (fig 2G ▶) ileum from CD patients. In addition, we found that two CD patients homozygous for the L1007fsinsC mutation expressed detectable NOD2 protein in Paneth cells (fig 2H, I ▶). The latter is consistent with the observation that 2D9 recognises the L1007fsinsC mutant protein (fig 1C ▶). Incubation with isotype matched mouse monoclonal antibody did not show any reactivity under our staining conditions (fig 2B ▶). The cellular pattern of NOD2 staining in the ileal epithelium was similar to that observed for lysozyme, a marker of Paneth cells (fig 2E ▶). However, in contrast with lysozyme which was primarily localised inside the granules (fig F), NOD2 reactivity was detected in the cytosol but in close proximity to the granules (fig 2D ▶).
Figure 2.
Expression of NOD2 in terminal ileum from Crohn’s disease (CD) patients and controls. (A–D) Uninvolved terminal ileum from a CD patient stained with 2D9 (A, C, D) or B-14 isotype matched control antibody (B). (E, F) Uninvolved ileum from a CD patient stained with antilysozyme antibody. (D) and (F) are high power views of Paneth cells from (A) and (E), respectively, illustrating NOD2 labelling at the periphery of secretory granules and lysozymes inside the granules. (G) Inflamed terminal ileum from a CD patient stained with 2D9, showing Paneth cell staining (indicated by arrowheads). (H, I) Medium power view of Paneth cells stained with 2D9 (arrowheads) from two different CD patients homozygous for the L1007fsinsC mutation. Magnification: 250× (A, B, E); 500× (C, G); 1000× (H, I); 2500× (D, F). The results shown in panels A–D, E, F, G, H, and I are from different patients and are representative of all patients and controls studied.
Ileal crypts are enriched for NOD2 mRNA expression
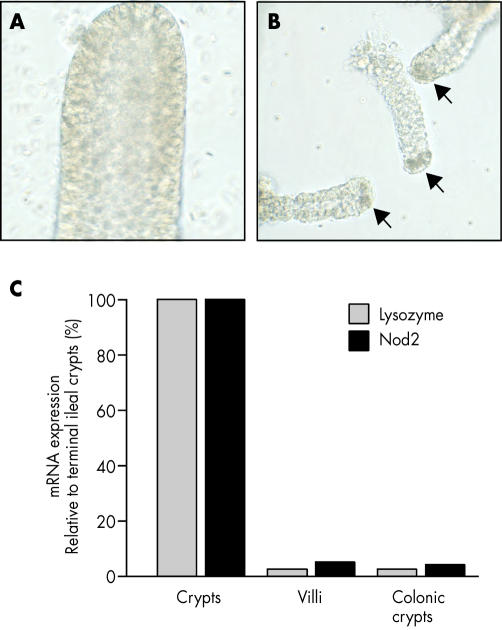
To further support the evidence that NOD2 is expressed in Paneth cells, we used calcium chelation and mechanical agitation to purify the ileal and colonic crypts and villous epithelial cells. Using this procedure we obtained highly enriched populations of ileal crypts with Paneth cells (fig 3A, B ▶). We used real time RT-PCR to quantitatively assess expression of NOD2 mRNA in crypt and villous cell fractions. The analysis revealed that ileal crypts expressed approximately 20-fold more NOD2 mRNA than colonic crypts (known to contain no or few Paneth cells) or villi enriched fractions (fig 3C ▶). Analysis of lysozyme mRNA in the same fractions showed that lysozyme expression was detected preferentially in ileal crypts, indicating that villi and colonic crypts were largely devoid of lysozyme expressing cells (fig 3C ▶).
Figure 3.
Expression of NOD2 mRNA in crypt and villus epithelial cells. (A) Whole villi and (B) crypts containing Paneth cells (arrows) were obtained by calcium chelation and mechanical shaking of mucosal tissue dissected from surgically resected small intestinal segments, and viewed under the 20× objective in an inverted phase microscope. (C) Quantitative real time reverse transcription-polymerase chain reaction (RT-PCR) analysis of crypt and villus epithelium shows enrichment of lysozyme and NOD2 mRNA expression in crypts. Lysozyme is approximately 40-fold enriched in crypts and NOD2 approximately 20-fold enriched in crypts from the terminal ileum compared with villi or colonic crypts. Relative expression of NOD2 and lysozyme mRNA was determined by real time RT-PCR, using intron spanning primers, and NOD2:glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and lysozyme:GAPDH ratios, calculated for each sample. Results are shown as relative expression compared with the level in small intestinal crypts.
NOD2 is expressed in metaplastic Paneth cells of the colon
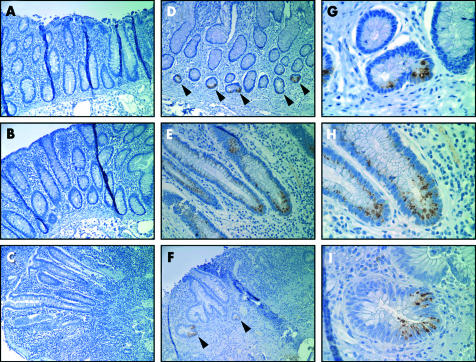
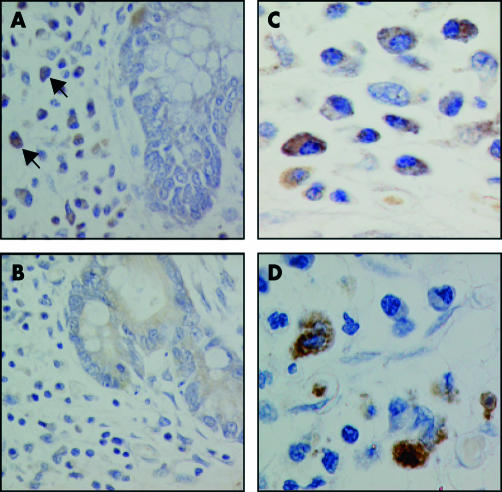
We also assessed expression of NOD2 in colon specimens. Staining of histologically normal colon specimens from five control individuals or histologically normal colon from CD patients revealed no detectable expression of NOD2 in the colonic epithelium (fig 4A ▶). Analysis of inflamed colon specimens from CD and UC patients showed no significant staining in the colonic epithelium (fig 4B, C ▶), although focal NOD2 staining was identified in two CD and one UC patient (fig 4E–I ▶). Notably, we found staining for NOD2 in colonic epithelium with Paneth cell metaplasia from a CD patient (fig 4D–G ▶) which is consistent with expression of NOD2 in Paneth cells. In addition, minimal or no staining for NOD2 was observed in the lamina propia or villous epithelium of uninflamed tissue in the ileum (fig 2A ▶, 2D ▶, 2H–I ▶) and colon (figure 4A ▶, 4G–I ▶). However, in colonic sections from patients with CD, infiltrating monocytes were stained using the anti-NOD2 antibody (fig 5 ▶). Lamina propia cells expressing NOD2 had indented kidney shaped nuclei, moderate amounts of cytoplasm, and expressed CD68 (fig 5 ▶). These morphological features together with CD68 labelling is characteristic of newly recruited monocytes. Mature resident tissue macrophages in the terminal which stained for CD68 but lacked the characteristic morphology of monocytes were not stained using the antibody, suggesting that expression of NOD2 is downregulated during monocyte differentiation.
Figure 4.
Expression of NOD2 in colon from Crohn’s disease (CD) patients and controls. (A, B) Uninvolved colon from a CD patient stained with 2D9 (A) or with isotype matched control antibody (B). (C) Inflamed colon from a CD patient showing no detectable 2D9 staining in the colonic epithelium. (D) Distal colon from a CD patient with Paneth cell metaplasia (indicated by arrowheads) stained with 2D9. (E) Inflamed colon from a CD patient showing focal NOD2 staining in colonic epithelium. (F) Inflamed colon from an ulcerative colitis patient showing focal NOD2 staining in colonic epithelium (indicated by arrowheads). (G–I) High power views of (D), (E), and (F), respectively. Magnification: 500× (A–F); 2500× (G–I).
Figure 5.
NOD2 protein expression in lamina propria mononuclear phagocytes in Crohn’s disease (CD). (A) NOD2 protein is detected within lamina propria mononuclear cells (arrows) in CD by immunohistochemical staining using the 2D9 antibody. (B) No positive staining is noted in parallel sections incubated with no primary antibody. (C) At higher magnification, NOD2 protein is detected in the cytoplasm of lamina propria cells. (D) Immunohistochemical staining using an anti-CD68 antibody verifies the identity of lamina propria monocytes. All sections were counterstained with haematoxylin and viewed under a 40× (A, B) or 100× (C, D) objective.
DISCUSSION
The discovery that NOD2 mutations confer susceptibility to CD has provided important insight into the pathogenesis of the disease. The finding that NOD2 can function as a sensor for bacterial components and can mediate NFκB activation suggests that this protein plays a role in host defence against microbes. NOD2 mutations are deficient in response to bacteria, supporting a model in which impaired recognition of luminal bacteria triggers an aberrant inflammatory response in the intestinal tissue of CD patients.13,14 However, NOD2 was found to be expressed predominantly in monocytes and other peripheral blood immune cells and this cellular distribution cannot explain fully the specific association of NOD2 mutations with ileal disease. In the present study, we developed a monoclonal antibody specific for human NOD2 to study expression of NOD2 in the ileum and colon of CD patients and controls. Our results show that NOD2 is expressed in Paneth cells of the small intestinal epithelium in both CD patients and controls. We observed that the villous and colonic epithelium was largely devoid of NOD2 staining, as detected with our monoclonal anti-NOD2 antibody. However, we found focal NOD2 staining in newly recruited monocytes/macrophages in the lamina propia of the colon from CD patients, which is consistent with expression of NOD2 in monocytes.8,16 In addition, NOD2 was detected in colonic epithelium in a subset of patients with inflammatory bowel disease at the site of inflammation and in metaplastic Paneth cells in the colon. Recent studies have shown that stimulation of intestinal epithelial cells with TNF-α upregulates NOD2 gene expression through transcriptional activation of NFκB sites in the NOD2 promoter.16,23,24 Thus it is likely that in some patients, TNF-α and/or other cytokines produced locally at inflammatory sites could play a role in induction of NOD2 in the intestinal epithelium. The significance of induced NOD2 in the colonic epithelium is unclear and awaits further investigation. NOD2 mRNA has been detected in unstimulated primary epithelial cells derived from colonic crypts by a non-quantitative RT-PCR assay.24 We did detect NOD2 mRNA in uninflammed ileal villi or colonic crypts by real time RT-PCR, although levels were very low compared with levels detected in ileal crypts.
Paneth cells play an important role in the protection of the host against luminal bacteria and have been implicated in the control of the enteric microflora.17,18 The mechanism by which Paneth cells sense bacteria in the gut is poorly understood but it may involve receptors for bacterial components expressed in Paneth cells or neighbouring cells. In this respect, secretion of antimicrobial products by Paneth cells is induced in response to bacterial products, including muramyl dipeptide, the molecule recognised by NOD2.19 However, the system by which Paneth cells recognise bacteria has not been identified. Expression of NOD2 in Paneth cells suggests that NOD2 could be part of a recognition apparatus for luminal bacteria. Because NOD2 mutations associated with CD are impaired in the response to bacterial components,5,13,14 it is reasonable to suggest that individuals harbouring NOD2 mutations could exhibit alteration in the secretion of antimicrobial peptide in response to gut microflora, defective clearing of luminal bacteria, and increased susceptibility to certain microbes. The recent observation that expression of β-defensin 1 is reduced in intestinal tissue from CD patients is significant in this regard.20 Such defects in Paneth cells could play a role in triggering the inflammatory response observed in CD patients.
Acknowledgments
This work was supported by grants R01 DK61707 and 5P60-DK20572 from the National Institutes of Health. YO was supported by a fellowship from the Crohn’s and Colitis Foundation of America.
Abbreviations
CD, Crohn’s disease
UC, ulcerative colitis
CARD, caspase recruitment domain
LPS, lipopolysaccharide
PGN, peptidoglycan
NFκB, nuclear factor κB
RT-PCR, reverse transcription-polymerase chain reaction
PBS, phosphate buffered saline
GAPDH, glyceraldehyde-3-phosphate dehydrogenase
TNF-α, tumour necrosis factor α
REFERENCES
- 1.Loftus E, Silverstein MD, Sandborn W, et al. Crohn’s disease in Olmsted County, Minnesota, 1940–1993: incidence, prevalence, and survival. Gastroenterology 1998;114:1161–8. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein CN, Blanchard JF, Rawsthorne P, et al. Epidemiology of Crohn’s disease and ulcerative colitis in a central Canadian province: a population-based study. Am J Epidemiol 1999;149:916–24. [DOI] [PubMed] [Google Scholar]
- 3.Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology 1998;115:182–205. [DOI] [PubMed] [Google Scholar]
- 4.Hendrickson BA, Gokhale R, Cho JH. Clinical aspects and pathophysiology of inflammatory bowel disease. Clin Microbiol Rev 2002;15:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001;411:603–6. [DOI] [PubMed] [Google Scholar]
- 6.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- 7.Hampe J, Cuthbert A, Croucher PJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 2001;357:1925–8. [DOI] [PubMed] [Google Scholar]
- 8.Ogura Y, Inohara N, Benito A, et al. NOD2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-B. J Biol Chem 2001;276:4812–18. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad T, Armuzzi A, Bunce M, et al. The molecular classification of the clinical manifestations of Crohn’s disease. Gastroenterology 2002;122:854–66. [DOI] [PubMed] [Google Scholar]
- 10.Cuthbert A, Fisher S, Croucher PJ, et al. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology 2002;122:867–74. [DOI] [PubMed] [Google Scholar]
- 11.Lesage S, Zouali H, Cezard JP, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet 2002;70:845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inohara N, Ogura Y, Chen FF, et al. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J Biol Chem 2001;276:2551–4. [DOI] [PubMed] [Google Scholar]
- 13.Bonen DK, Ogura Y, Nicolae DL, et al. Crohn’s disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology 2003;124:140–6. [DOI] [PubMed] [Google Scholar]
- 14.Inohara N, Ogura Y, Fontalba A, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2: implications for Crohn’s disease. J Biol Chem 2003;278:5509–12. [DOI] [PubMed] [Google Scholar]
- 15.Girardin SE, Boneca IG, Viala J, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 2003;278:8869–72. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez O, Pipaon C, Inohara N, et al. Induction of NOD2 in myelomonocytic and intestinal epithelial cells via nuclear factor-kappa B activation. J Biol Chem 2002;277:41701–5. [DOI] [PubMed] [Google Scholar]
- 17.Ouellette AJ, Bevins CL. Paneth cell defensins and innate immunity of the small bowel. Inflamm Bowel Dis 2001;7:43–50. [DOI] [PubMed] [Google Scholar]
- 18.Wilson CL, Ouellette AJ, Satchell DP, et al. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 1999;286:113–17. [DOI] [PubMed] [Google Scholar]
- 19.Ayabe T, Satchell DP, Wilson CL, et al. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nature Immunol 2000;1:113–18. [DOI] [PubMed] [Google Scholar]
- 20.Wehkamp J, Fellermann KF, Herrlinger KR, et al. Human b-defensin 2 but not b-defensin 1 is expressed preferentially in colonic mucosa of inflammatory bowel disease. Eur J Gastroenterol Hepatol 2002;14:745–52. [DOI] [PubMed] [Google Scholar]
- 21.Kearney JF, Radbruch A, Liesegang B, et al. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol 1979;123:1548–50. [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibody—a laboratory manual. New York: Cold Spring Harbor Laboratory, 1988:283–318.
- 23.Rosenstiel P, Fantini M, Brautigam K, et al. TNF-alpha and IFN-gamma regulate the expression of the NOD2 (CARD15) gene in human intestinal epithelial cells. Gastroenterology 2003;124:1001–9. [DOI] [PubMed] [Google Scholar]
- 24.Hisamatsu T, Suzuki M, Reinecker HC, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology 2003;124:993–1000. [DOI] [PubMed] [Google Scholar]