Abstract
Spore formation in Saccharomyces cerevisiae requires the synthesis of prospore membranes (PSMs) followed by the assembly of spore walls (SWs). We have characterized extensively the phenotypes of mutants in the sporulation-specific genes, SSP2 and OSW1, which are required for spore formation. A striking feature of the osw1 phenotype is asynchrony of spore development, with some spores displaying defects in PSM formation and others spores in the same ascus blocked at various stages in SW development. The Osw1 protein localizes to spindle pole bodies (SPBs) during meiotic nuclear division and subsequently to PSMs/SWs. We propose that Osw1 performs a regulatory function required to coordinate the different stages of spore morphogenesis. In the ssp2 mutant, nuclei are surrounded by PSMs and SWs; however, PSMs and SWs often also encapsulate anucleate bodies both inside and outside of spores. In addition, the SW is not as thick as in wild type. The ssp2 mutant defect is partially suppressed by overproduction of either Spo14 or Sso1, both of which promote the fusion of vesicles at the outer plaque of the SPB early in PSM formation. We propose that Ssp2 plays a role in vesicle fusion during PSM formation.
SPORULATION in the budding yeast, Saccharomyces cerevisiae, involves two overlapping processes—meiosis and spore morphogenesis. During meiosis, a diploid cell undergoes a single round of DNA replication, followed by two rounds of chromosome segregation, to generate four haploid nuclei. During spore morphogenesis, the four daughter nuclei are packaged into spores, which are contained within an ascus.
Spore morphogenesis (reviewed by Neiman 2005) initiates with enlargement of the outer surface of the spindle pole body (SPB) to form the meiotic outer plaque. As cells undergo meiosis II, vesicles coalesce on the meiotic plaque to form a novel intracellular membrane called the prospore membrane (PSM). The double-layered PSM extends outward from each SPB to surround the adjacent nucleus. When the leading edges of the PSM meet and become fused, the haploid nucleus is fully encapsulated to form a prospore. PSM closure triggers the final step of spore morphogenesis, spore wall (SW) formation. The SW is assembled between the two membranes of the PSM and consists of four layers. The two inner layers are composed of glucan and mannan, which are components of vegetative cell walls. The two outer layers are composed of chitosan and dityrosine and provide much of the spore's resistance to environmental stress. The inner layer of the former PSM forms the plasma membrane underlying the SW; the outer layer of the PSM breaks down during SW formation.
Genetic analysis indicates that the vesicles that fuse to form the PSM are post-Golgi vesicles, closely related to those normally involved in secretion (Neiman 1998). A widely accepted model for vesicle fusion events in the secretory pathway is the soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) hypothesis. According to this model, every vesicle carries on its surface a vesicular (v-)SNARE that can interact only with a target (t-)SNARE on the correct acceptor membrane (Jahn and Sudhof 1999). In the case of secretion in budding yeast, vesicles destined for the plasma membrane carry on their surface the v-SNARE molecules, Snc1 and Snc2. The Snc proteins interact specifically with the t-SNARE proteins, Sec9, and Sso1 and/or Sso2, on the plasma membrane (Gerst 1999).
The synthesis of PSMs during sporulation also depends on SNAREs, although the players differ somewhat from those used during secretion in vegetative cells. In vegetative cells, Sso1 and Sso2 function redundantly. Sporulation, however, specifically requires Sso1 (but not Sso2) (Jantti et al. 2002; Enyenihi and Saunders 2003). Sec9 is essential for secretion in vegetative cells, but it plays only a minor role during sporulation, where its function is largely replaced by the sporulation-specific Sec9 homolog, Spo20 (Neiman 1998). In the sso1 mutant or in the sec9 spo20 double mutant, secretory vesicles accumulate near the meiotic plaque, but fail to coalesce to form a membrane sheet (Nakanishi et al. 2006).
Another protein important for PSM formation is Spo14, a phospholipase D enzyme. Spo14 is essential during sporulation, and it localizes to PSMs (Rose et al. 1995). The phenotype of a spo14 null mutant is similar to that of the sso1 and sec9 spo20 mutants, indicating that Spo14 also participates in vesicle fusion (Riedel et al. 2005; Nakanishi et al. 2006). Studies of nonnull alleles of SPO14 and of mutations affecting the Spo14-interacting protein, Sma1, indicate that Spo14 acts at two discrete steps in PSM formation: the initial fusion of vesicles on the meiotic plaque and subsequent enlargement of the PSM to surround the adjoining nucleus (Riedel et al. 2005; Nakanishi et al. 2006).
Expansion of the PSM depends on a complex of proteins, called the leading-edge complex, which includes Don1, Ssp1, and Ady3 (Moreno-Borchart et al. 2001; Nickas and Neiman 2002). These proteins initially localize to the SPB; as the PSM expands to engulf the adjacent nucleus, they localize to a ring corresponding to the growing edge of the PSM. The leading-edge complex is believed to be important in determining the shape and size of the PSM and in regulating the amount of cytoplasm surrounding each encapsulated nucleus (Neiman 2005).
In a screen for genes required for sporulation, we identified the sporulation-specific (SSP)2 and outer spore wall (OSW)1 genes. Concurrent with our studies, SSP2 and OSW1 were also characterized by two other groups (Sarkar et al. 2002; Coluccio et al. 2004). Our results indicate that the ssp2 and osw1 null mutants are proficient in meiotic recombination and chromosome segregation, but defective in the subsequent processes of PSM and SW formation. The osw1 mutant exhibits a heterogeneous phenotype with different spores in the same ascus arrested at different stages of PSM and SW formation. On the basis of the similarity of osw1 to various kinase mutants, we suggest that the Osw1 protein plays a role in regulating and coordinating spore morphogenesis. In the ssp2 mutant, nuclei are surrounded by PSMs and SWs, but the SW is only half the thickness of its wild-type counterpart. In addition, anucleate spore-like bodies are generated, raising the possibility that PSM formation is not tightly coupled to SPBs. The ssp2 defect is partially suppressed by overproduction of Spo14 or Sso1, suggesting that Ssp2 plays a role in vesicle fusion during PSM formation.
MATERIALS AND METHODS
Plasmids:
Standard methods were used in plasmid constructions (Sambrook et al. 1989). To construct an SSP2 disruption, SSP2 was first amplified by PCR from the S. cerevisiae λ-clone 6767 (ATCC 70786) to introduce a BamHI site ∼270 bp upstream of the SSP2 ORF and a KpnI site ∼80 bp downstream. The BamHI–KpnI fragment containing SSP2 was inserted between the BamHI and KpnI sites of Bluescript SK (+) (Stratagene, La Jolla, CA) to create pJS1. To construct pJS2 (ssp2∷URA3), the ∼1-kbp HindIII–SalI fragment of URA3 (Chua and Roeder 1998) was used to replace the HindIII–XhoI fragment of SSP2 (nucleotides 71–727 of the coding region) in pJS1.
An SSP2–GFP fusion gene was constructed as follows. PCR was used to introduce a NotI site ∼250 bp upstream of the SSP2 ORF and an NheI site just before the stop codon. The NotI–NheI fragment (∼1.4 kbp) containing SSP2 was then subcloned into the NotI–NheI sites of pSA111 (Agarwal and Roeder 2000), thus creating pJ82, which contains the coding sequences for green fluorescent protein (GFP) fused to the 3′ end of SSP2.
To construct an OSW1 disruption, a 2.0-kbp EcoRI fragment containing OSW1 from the S. cerevisiae λ clone 6933 (ATCC 71157) was first subcloned into pHSS6 (Seifert et al. 1986) to create pJS4. To construct pJS5 (osw1∷URA3), the ∼1-kbp HindIII–SmaI fragment of URA3 (Chua and Roeder 1998) was then used to replace the HincII–HindIII fragment of OSW1 (nucleotides 70–721 of the coding region) in pJS4.
An OSW1–GFP fusion gene was constructed as follows. PCR was used to introduce a NotI site ∼290 bp upstream of the OSW1 ORF and an NheI site just before the stop codon. The NotI–NheI fragment (∼1.1 kbp) containing OSW1 was subcloned into the NotI–NheI sites of pSA111 (Agarwal and Roeder 2000) to create pJ83, which contains GFP fused to the 3′ end of OSW1.
pSB8 is a centromere-containing, pRS314-based plasmid encoding Don1–GFP (Tachikawa et al. 2001). pME1086 is a multicopy YEp351-based plasmid encoding Spo14–GFP (Rudge et al. 1998). The XbaI–ScaI fragment (∼4.5 kbp) containing part of SPO14 and the full sequence of GFP from the SPO14–GFP plasmid was subcloned into the XbaI–SmaI sites of the integrating plasmid pRS305 (Sikorski and Hieter 1989) to create pJ121. YEpSSO1 and YEpSSO2 are pMAC561-based plasmids containing the SSO1 and SSO2 genes (Ruohonen et al. 1997); pMA56 was used as the control plasmid.
Yeast strains:
Yeast manipulations were performed, and media were prepared as described previously (Sherman et al. 1986). Gene disruptions were confirmed by PCR analysis. All strains used are isogenic with BR2495 (Rockmill and Roeder 1990), which has the following genotype: MATa/MATα his4-280/his4-260 leu2-27/leu2-3,112 arg4-8/ARG4 thr1-1/thr1-4 cyh10/CYH10 ade2-1/ade2-1 ura3-1/ura3-1 trp1-1/trp1-289. Diploids used in this study were made by mating appropriate haploids, generated by transformation and/or genetic crosses. All strains used are diploids homozygous for the disruptions/fusions indicated.
ssp2∷URA3 and osw1∷URA3 mutant strains were constructed by transforming with the BamHI–KpnI fragment of pJS2 and the EcoRI fragment of pJS5, respectively.
pAFS59 (Straight et al. 1996) containing 256 Lac operator (LacO) repeats was used to integrate LacO at the LEU2 locus, which is located 22 kbp to the left of CENIII. pAFS152 (Shonn et al. 2000), containing the GFP–Lac repressor (LacI) fusion driven by the CYC1 promoter, was integrated at the URA3 locus.
pJ121 was digested with BsaBI and then used to transform wild-type, ssp2∷URA3 and osw1∷URA3 haploids, creating strains in which SPO14–GFP is integrated at its genomic loci.
Measurements of sporulation and nuclear division:
To measure sporulation efficiency, each saturated culture (1 ml) was washed once with distilled water and resuspended in 10 ml of sporulation medium (2% potassium acetate). After 72 hr at 30°, cells were observed by differential interference contrast (DIC) microscopy. Three cultures were examined for each strain, and at least 100 cells were counted for each culture. The percentages presented are averages.
To determine the frequency and kinetics of meiotic nuclear division, cells were fixed in 70% ethanol and frozen at −20° prior to staining with 4′-6-diamidino-2-phenylindole (DAPI) (Hong and Roeder 2002). Cells were observed and counted under the fluorescence microscope (Nikon Eclipse E800, Plan Apo100×/1.4 oil objective).
Chromosome segregation was monitored in wild-type, ssp2∷URA3, and osw1∷URA3 strains with the LacO-marked chromosome III (LEU2) bound with GFP–LacI (Straight et al. 1996). The cells were first fixed with formaldehyde, stained with DAPI (1 μg/ml), and then observed under the fluorescence microscope. All fluorescence images presented in this article were captured with a fluorescence microscope (Nikon Eclipse E800, Plan Apo100×/1.4 oil objective) equipped with an FITC HYQ filter to visualize the GFP signals. Images were captured with a Sensys CCD camera (Photometrics, Tucson, AZ). Each image presented represents a single focal plane.
Analysis of spread nuclei:
Meiotic nuclei were spread and stained with antibodies as described (Bailis and Roeder 1998). Guinea pig anti-GFP antibodies (Tsubouchi and Roeder 2002) and rat anti-tubulin antibodies (Sera-Lab) were used at a 1:100 dilution. Anti-rabbit antibodies conjugated with FITC and anti-rat antibodies conjugated with Texas Red (both from Jackson ImmunoResearch Labs, West Grove, PA) were used at a 1:200 dilution as secondary antibodies. Chromosomal DNA was stained with 1 μg/ml DAPI.
Staining of fixed whole cells:
To observe the Ssp2–GFP fluorescent signal combined with DAPI staining in whole cells, ∼20 μl of sporulating cells were scraped from sporulation plates and suspended in solution A that contains 3.7% formaldehyde, 1.2 m sorbitol, and 50 mm potassium phosphate buffer, pH 6.8. After fixation for 1 hr at room temperature with rotation, cells were washed three times with solution A and incubated at 37° for 1 hr in solution A supplemented with 0.1% β-mercaptoethanol and 25 μg/ml zymolyase. Cells were washed twice with solution A and resuspended in a small volume of phosphate-buffered saline (PBS). Cells were then added to Poly-l-lysine coated slides and rinsed once with PBS followed by PBS + 0.1% NP-40. Guinea pig anti-GFP antibody (Tsubouchi and Roeder 2002) was diluted 1:100 in PBS + 1% BSA and used to stain cells. After overnight incubation at 4°, cells were rinsed with PBS, then PBS + 0.1% NP-40, and then PBS. Anti-guinea pig antibodies conjugated with FITC and diluted 1:200 in PBS + 1% BSA were used as secondary antibodies. After 2.5 hr of incubation at room temperature, the cells were rinsed in PBS as described above and then mounted with DAPI.
Electron microscopy:
Samples for examination in the electron microscope (EM) were prepared by potassium permanganate (KMnO4) fixation as described by Du and Novick (2002). Samples were sectioned using a Sorvall MT-2 Ultramicrotome (Waltham, MA) and then analyzed using a Zeiss EM 10A transmission EM (Zeiss, Jena, Germany). For measurements of SW thickness, 11 asci were examined for wild type and 13 for ssp2.
RESULTS
Identification of the SSP2 and OSW1 genes:
We previously identified transposon insertions that generate in-frame lacZ fusion genes expressed specifically during sporulation (Burns et al. 1994; Ross-Macdonald et al. 1999). Diploids homozygous for these insertions were constructed and screened for sporulation competence. Diploids derived from strains M86 and M88 were unable to sporulate; these represent insertions in ORFs YOR242c and YOR255w, respectively. Both of these ORFs were identified in previous screens for sporulation-defective mutants (Rabitsch et al. 2001; Sarkar et al. 2002; Enyenihi and Saunders 2003). YOR242c has been termed SSP2 by Sarkar et al. (2002), and YOR255w has been named OSW1 by Coluccio et al. (2004). SSP2 encodes a protein of 371 amino acids, which shares significant homology with IPF16670, a predicted gene product of Candida albicans. OSW1 encodes a protein of 278 amino acids with no significant similarity to other proteins in databases.
Ssp2 and Osw1 are required for sporulation:
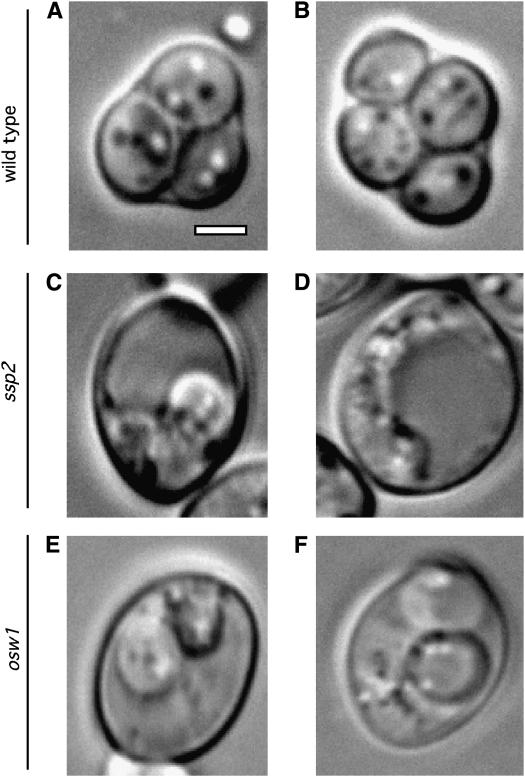
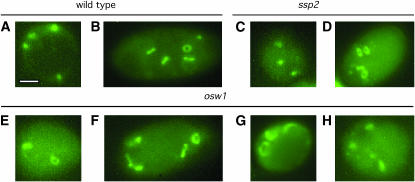
Deletion/disruption mutants of SSP2 and OSW1 were made by replacing most of each coding region with the URA3 gene (see materials and methods). Homozygous ssp2 and osw1 diploids and an isogenic wild type were tested for sporulation efficiency by DIC microscopy (see materials and methods). In wild type, 65% of cells produce mature four-spored asci (Figure 1, A and B) after 72 hr in sporulation medium. However, only 5 and 1% of cells generate mature asci in ssp2 and osw1, respectively. A significant fraction of cells (63% in ssp2 and 57% in osw1) appear to contain compartments, which are irregular in size and shape, suggestive of immature spores (Figure 1, C–F). The Ssp2 and Osw1 proteins are therefore required for spore formation.
Figure 1.—
Ssp2 and Osw1 are essential for sporulation. Typical cells of wild type (A and B), ssp2 (C and D), and osw1 (E and F) are shown. Cells were sporulated and observed under DIC microscopy after 72 hr in sporulation medium. Bar, 2 μm.
Meiotic division is not affected in ssp2 and osw1 mutants:
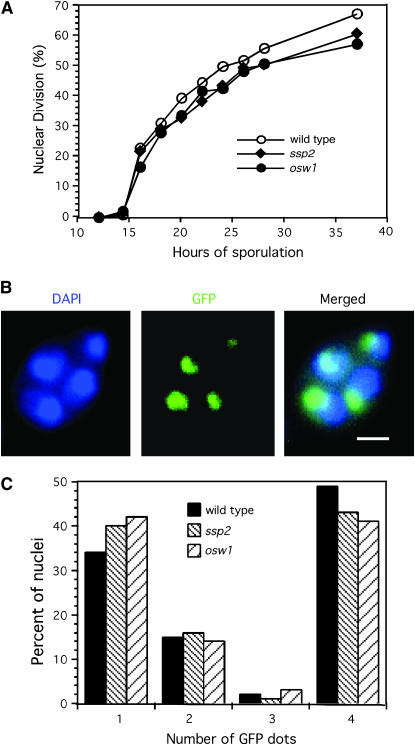
To assess meiotic nuclear division, cells harvested at different time points after the induction of meiosis were examined by fluorescence microscopy after staining with DAPI, a DNA-binding dye. Cells that have completed the meiosis I division contain two discrete nuclei, whereas cells that have completed both meiotic divisions contain four nuclei. Wild-type, ssp2, and osw1 cells undergo nuclear division with nearly identical kinetics (Figure 2A). There is no evidence of nuclear fragmentation in the mutants; four DAPI-stained bodies, equivalent in size, are present in nearly every ascus (data not shown). Thus, analysis of nuclear division suggests that the kinetics and efficiency of meiotic progression are normal in the ssp2 and osw1 mutants.
Figure 2.—
Chromosome segregation occurs normally in ssp2 and osw1. (A) Meiotic nuclear division was examined in wild-type, ssp2∷URA3, and osw1∷URA3 strains. Both binucleate and tetranucleate cells were scored as having undergone nuclear division. (B and C) Chromosome segregation was monitored in a strain that produces GFP–LacI fusion protein and is homozygous for a tandem array of LacO repeats inserted 22 kbp from CENIII. (B) An example of a cell that has undergone normal segregation is shown. Four DAPI-stained bodies indicate that chromosomes have undergone both meiotic divisions. One GFP dot per DAPI-stained body indicates that GFP-tagged chromosomes have segregated correctly. Bar, 2 μm. (C) Quantitation of GFP signals in wild-type, ssp2∷URA3, and osw1∷URA3 strains. Cells were harvested after 24 hr of sporulation, stained with DAPI, and examined by fluorescence microscopy. The percentages of cells with one, two, three, or four GFP dots are indicated. At least 100 cells were scored for each strain. The experiments shown in A and C were performed twice with qualitatively similar results.
Commitment to meiotic recombination is normal in the ssp2 and osw1 mutants:
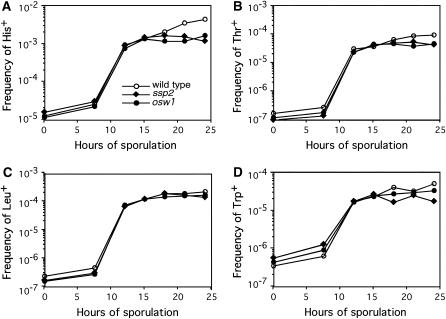
To examine the effect of ssp2 and osw1 on meiotic recombination, the null mutants were used to measure commitment to gene conversion (intragenic recombination) in return-to-growth assays. Cells were introduced into sporulation medium and then returned to growth medium prior to the commitment to meiotic chromosome segregation (Esposito and Esposito 1974). The frequency of gene conversion was measured in ssp2, osw1, and an isogenic wild type by selecting for prototrophic recombinants at four different loci carrying heteroalleles conferring auxotrophy. Commitment to meiotic gene conversion is normal in ssp2 and osw1 mutants (Figure 3), with respect both to timing and to frequency.
Figure 3.—
Commitment to meiotic recombination is normal in ssp2 and osw1. (A–D) Intragenic recombination was measured in wild-type, ssp2∷URA3, and osw1∷URA3 strains sporulated at 30°. Prototroph formation was monitored at four different heteroallelic loci at different time points during sporulation as described by Sym et al. (1993). The frequencies of prototroph formation at HIS4 (his4-280/his4-260), THR1 (thr1-1/thr1-4), LEU2 (leu2-27/leu2-3,112), and TRP1 (trp1-1/trp1-289) are shown as a function of time.
Chromosome segregation is not affected in the ssp2 and osw1 mutants:
To examine chromosome segregation, strains were constructed that produce a GFP–LacI fusion protein and are homozygous for a tandem array of LacO repeats inserted 22 kbp from CENIII (Straight et al. 1996) (see materials and methods). Thus, segregation of the LacO repeats, and by implication the centromere of chromosome III, can be monitored by fluorescence microscopic detection of the GFP–LacI protein (Straight et al. 1996).
Wild-type, ssp2, and osw1 cells were harvested after 24 hr of sporulation, when ∼60% of wild-type cells had undergone one or both nuclear divisions (Figure 2A). Cells were fixed, stained with DAPI, and observed by fluorescence microscopy. In wild type, some cells show only one GFP signal, indicative of meiotic prophase (or a failure to enter meiosis). Others show two distinct GFP signals, indicative of meiosis I division products. Some show four distinct GFP dots (one per DAPI-stained body), indicating the end of meiosis II (Figure 2B). The ssp2 and osw1 mutants show the same pattern of GFP signals as wild type (Figure 2C), indicating that chromosomes segregate correctly in the absence of Ssp2 or Osw1. Since crossing over is required for correct chromosome segregation in meiosis, these results indicate that the ssp2 and osw1 mutants are proficient in crossing over.
Localization of the Ssp2 and Osw1 proteins during meiosis:
SSP2–GFP and OSW1–GFP fusion genes were constructed (see materials and methods) and used to determine the intracellular locations of Ssp2 and Osw1. Single-copy plasmids carrying the fusion genes do not complement. However, when the fusion genes are present on a multicopy plasmid (carrying the 2μ circle origin of DNA replication), they enable the corresponding null mutants to sporulate as efficiently as wild type, indicating that the fusion proteins are partially functional. The ssp2 + 2μ SSP2–GFP and osw1 + 2μ OSW1–GFP strains were introduced into sporulation medium, and cells harvested at various time points were observed by fluorescence microscopy.
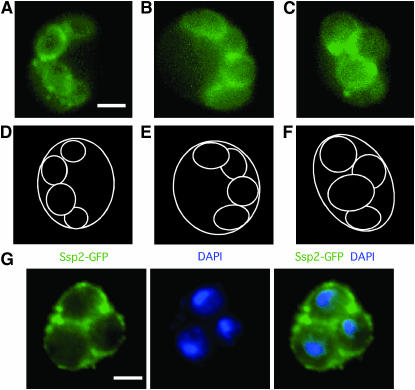
Figure 4 shows images of living cells expressing SSP2–GFP during the course of sporulation. In vegetative cells and early in meiosis, no Ssp2–GFP signal is detected (data not shown). Starting around 18 hr, when many cells have completed the meiosis II division, the Ssp2–GFP signal is detected as small ring-like structures (Figure 4, A and B). By 25 hr, the rings in most cells have enlarged to occupy most of the ascus (Figure 4C). Each Ssp2–GFP ring surrounds an individual nucleus, as determined by staining fixed cells both with anti-GFP antibodies and with DAPI (Figure 4G). Thus, the Ssp2–GFP signals observed must be associated with PSMs and/or SWs. Sarkar et al. (2002) also reported Ssp2 localization to rings surrounding nuclei starting after the meiosis II division.
Figure 4.—
Localization of Ssp2–GFP in sporulating cells. (A–F) The ssp2 mutant producing Ssp2–GFP was sporulated and then analyzed by observing GFP fluorescence in living cells. (A–C) Representative cells producing Ssp2–GFP. (D–F) Diagrams of the images in A–C. The diagram is based on the background GFP signal a cell displays when overexposed. Bar, 2 μm. (G) ssp2 cells containing 2μ SSP2–GFP were fixed and then stained with DAPI (blue) and anti-GFP antibodies (green). The fourth DAPI-stained body is visible in a different plane of focus. Time points for each image: A–B, 20 hr; C, 24 hr; G, 24 hr.
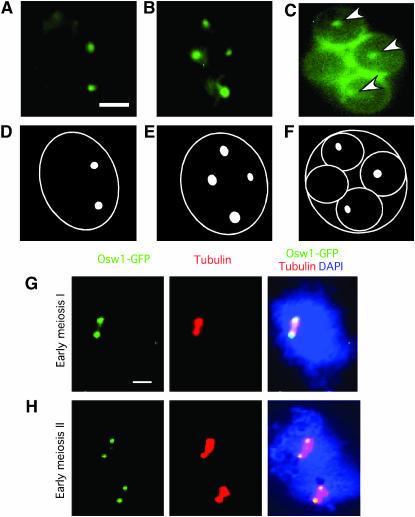
Osw1 localization shows different patterns at different times during sporulation. When living cells expressing OSW1–GFP are observed by fluorescence microscopy, Osw1–GFP first appears as two dots (Figure 5A) and later as four distinct foci (Figure 5B). Double staining of spread nuclei with anti-tubulin and anti-GFP antibodies indicates that these dots represent SPBs at meiosis I (two dots) and meiosis II (four dots) (Figure 5, G and H). After meiotic nuclear division is complete, the Osw1–GFP signals enlarge to fully surround nuclei, in addition to being present at the four SPBs (Figure 5C).
Figure 5.—
Localization of Osw1–GFP in sporulating cells. (A–F) The osw1 mutant producing Osw1–GFP was sporulated and then analyzed by observing GFP fluorescence in living cells. (A–C) Representative cells producing Osw1–GFP. (D–F) Diagrams of the images in A–C. In the case of the cell shown in C, three SPBs are evident (arrowheads), while the fourth SPB is evident in a different plane of focus. (G–H) Spread nuclei from osw1 containing 2μ OSW1–GFP were stained with DAPI (blue), anti-GFP (green), and anti-tubulin (red) antibodies. Bar, 2 μm. Time points for each image: A, 17 hr; B, 18 hr; C, 23 hr; G–H, 24 hr.
The leading edge of PSMs is affected by osw1 but not by ssp2:
Don1 was used as a marker to determine whether ssp2 and/or osw1 are defective in formation of the leading edge of the PSM. Living cells producing a Don1–GFP fusion protein were examined.
In wild type, Don1–GFP first appears as four distinct dots (Figure 6A). As the leading edge of the PSM moves away from the SPB to engulf the adjacent nucleus, Don1 staining appears as rings or bars (Figure 6B) depending on whether the leading edge of the membrane is viewed from the side (bars) or in cross section (rings). No obvious differences in Don1 localization were observed between the ssp2 mutant (Figure 6, C and D) and wild type.
Figure 6.—
Don1 rings are normal in ssp2, but aberrant in osw1. Wild-type (A and B), ssp2 (C and D), and osw1 (E–H) strains carrying DON1–GFP on a centromeric plasmid were sporulated. They were then analyzed by observing GFP fluorescence in living cells. The cell in E has two Don1 rings. In F, the top left Don1 signal is discontinuous, suggestive of incomplete ring formation. In H, the lower Don1 bar is significantly longer than the others. Time points for each image: A, 21 hr; B, 25 hr; C, 21 hr; D, 25 hr; E, 21 hr; F, 23 hr; G, 23 hr; H, 25 hr. Bar, 2 μm.
In the osw1 mutant, a variable number of Don1 dots/rings (one to four) were observed (Figure 6, E–H). Some cells contain partially developed or aberrantly shaped Don1 rings (Figure 6G); other cells contain four Don1 dots or rings at different stages of development (Figure 6, F and H). These observations suggest that Don1 rings, and thus the leading edges of the PSMs, develop aberrantly and asynchronously in the absence of Osw1.
Both Ssp2 and Osw1 are required for proper localization of Spo14:
To examine PSM formation further, the Spo14 protein, a marker for the PSM, was localized in diploid cells in which both copies of the SPO14 gene had been disrupted by insertion of a SPO14–GFP fusion gene (see materials and methods). SPO14–GFP integration has no effect on sporulation efficiency in wild-type, ssp2, or osw1 strains (data not shown). No specific GFP signal was observed in living cells. Thus, to detect Spo14–GFP, cells were fixed and stained with anti-GFP antibodies; cells were simultaneously stained with DAPI to visualize nuclei.
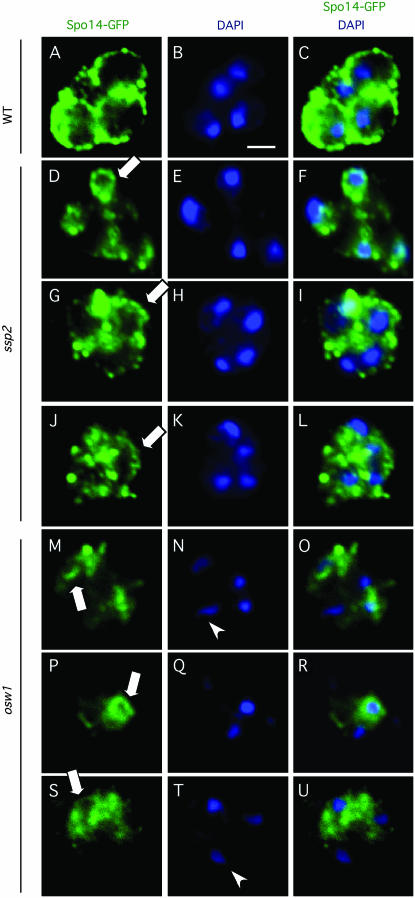
When sporulation is complete, ∼70% of wild-type cells show four Spo14–GFP circles around four DAPI-stained nuclear bodies (Figure 7, A–C). However, none of the ssp2 cells show a similar pattern. Instead, Spo14–GFP signals most often appear as punctate dots dispersed around DAPI signals (Figure 7, D–L). ssp2 cells sometimes display linear stretches of continuous Spo14 staining (Figure 7, G and J, open arrows); occasionally, small Spo14–GFP circles centered around DAPI-stained material are observed (Figure 7D, open arrow). Large circles surrounded by Spo14–GFP staining (like the circles seen in wild type) were never observed. The predominantly punctate staining pattern and the absence of large circles suggest that PSMs do not mature properly in the ssp2 mutant.
Figure 7.—
The localization of Spo14–GFP is aberrant in ssp2 and osw1. Wild-type, ssp2, and osw1 strains with SPO14–GFP integrated (see materials and methods) were sporulated for 3 days before cells were fixed and stained with DAPI (blue) and rabbit anti-GFP antibodies (green). Arrows, small Spo14–GFP circles or short linear stretches of Spo14–GFP signals. Arrowheads, DAPI-stained bodies without surrounding Spo14–GFP signals. Bar, 2 μm. The fourth Spo14–GFP circle in A and C and the fourth DAPI-stained body in T and U were evident in another focus plane. No specific GFP signals were observed in a control strain lacking Spo14–GFP (data not shown).
Spo14–GFP mislocalizes even more severely in the osw1 mutant. Spo14–GFP staining often appears to be concentrated to one side of the ascus. There are no obvious circles of Spo14 staining, although short linear stretches are sometimes observed (Figure 7, M, P, and S, open arrows). DAPI-stained masses without any surrounding Spo14–GFP signal are often observed (Figure 7, N and T, arrowheads), suggesting the absence of a PSM.
Thus, Spo14–GFP mislocalizes in both mutants, indicating roles for Ssp2 and Osw1 in the formation of PSMs.
ssp2 and osw1 mutants form aberrant structures in prospores:
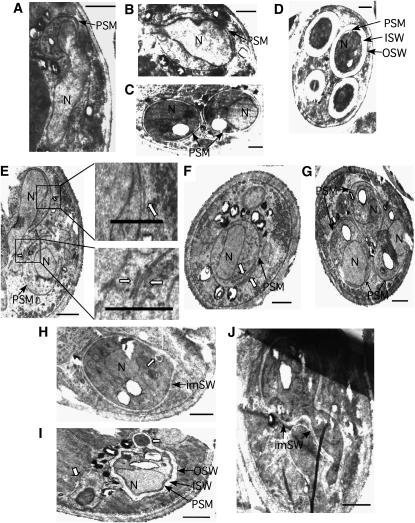
To investigate further the ssp2 and osw1 defects in spore formation, sectioned cells were examined in the EM. In wild-type cells, each PSM starts to develop at one side of a nucleus (Figure 8A), then expands to engulf the nucleus (Figure 8B), and eventually closes when the nucleus is fully surrounded. Later in spore morphogenesis, the SW materials are deposited between the two layers of the PSM (Figure 8C). In our EM preparations, the two innermost layers of the SW (composed of glucan and mannan) appear as one thick electron-translucent layer (Figure 8D), referred to hereafter as the inner SW (ISW). The two outer layers of the SW (composed of dityrosine and chitosan) appear as one thin darkly stained layer, referred to hereafter as the outer spore wall (OSW). The inner layer of the former PSM defines the plasma membrane underlying the mature SW.
Figure 8.—
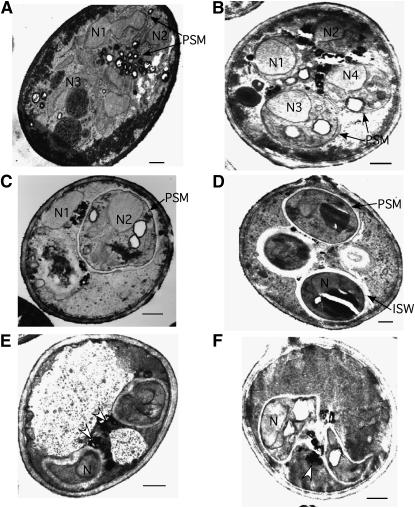
ssp2 forms aberrant PSMs and SWs. Wild-type (A–D) and ssp2 (E–J) strains were incubated in sporulation medium at 30°. (A and E) Cells were harvested at 24 hr, when ∼60% of cells have undergone one or both nuclear divisions. (B–D and F–J) Cells were harvested at 36 hr, when spore morphogenesis is complete in wild type. Arrows in E–G indicate aberrant membrane-like structures; arrows in H and I indicate aberrant anucleated spore-like bodies. N, nucleus; PSM, prospore membrane; PSM, prospore membrane; ISW, inner spore wall; OSW, outer spore wall; imSW, immature spore wall. Bar, 500 nm.
During the early stage of PSM development in the ssp2 mutant, PSMs are formed; however, short pieces of membrane-like structures are often seen close by the more fully developed PSMs (Figure 8E, arrows). The frequency of these membranous structures is 1.7 ± 0.9/spore section. (Similar pieces of membrane were never observed in wild type.) By the time the PSM has successfully encapsulated an entire daughter nucleus, longer stretches of free membrane are frequently observed (Figure 8, F and G, arrows). Prospores do form and SWs start to develop in the ssp2 mutant; however, PSMs and SWs often encapsulate unidentified electron-dense materials both inside and outside of spores (Figure 8, H and I, arrows). In addition, some spores are very irregular in shape (Figure 8J). The ISW does not reach the thickness seen in wild type (compare Figure 8, H–J, with 8D). The ISW is 0.22 ± 0.01 μm thick in wild type compared to 0.09 ± 0.01 μm in ssp2. The OSW appears to form normally in ssp2.
The osw1 mutant displays various defects. The most obvious is the asynchronous development of the four spores within an ascus. At the stage when PSMs develop, sometimes only a subset of nuclei become surrounded by PSMs (Figure 9, A–C). At the SW maturation stage, some asci contain prospores with immature SWs of different thicknesses (Figure 9D). A second defect is that the OSW is rarely seen (Figure 9, C–F). In addition, electron-dense materials often accumulate around the surface of the SW (Figure 9, E and F, arrowheads). This accumulation has been observed in other SW-defective mutants (Wagner et al. 1997; Christodoulidou et al. 1999; Tachikawa et al. 2001). Last, the immature spores sometimes are very irregular in shape (Figure 9, E and F), perhaps due to the absence of the OSW for stabilization.
Figure 9.—
osw1 forms aberrant PSMs and SWs. osw1 cells were analyzed as described. Open arrowheads in E and F, electron-dense materials around the surface of the SW. N, nucleus (numbered in A–C); PSM, prospore membrane; ISW, inner spore wall. (A) N1 and N2 are being engulfed by a PSM, while N3 does not have any PSM. (B) N1, N3, and N4 have PSMs, while N2 does not. (C) N2 is fully encapsulated by a PSM and an incipient SW structure is discernible, while N1 does not show any PSM formation. Bar, 500 nm.
Overproduction of Spo14 partially suppresses the sporulation defect of ssp2:
In the process of immunolocalizing the Spo14 protein, we discovered that overproduction of the Spo14–GFP protein partially suppresses the sporulation defect of the ssp2 mutant. When SPO14–GFP was introduced on a multicopy plasmid, 21% of ssp2 cells formed mature four-spored asci (as assessed by DIC microscopy), compared to only 5% of cells carrying an empty vector. Overproduction of Spo14–GFP does not alter the sporulation efficiency of wild type or osw1 (Rudge et al. 1998) (data not shown).
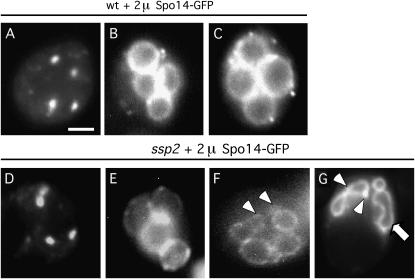
Both wild-type and ssp2 strains overproducing Spo14–GFP were sporulated, and living cells were examined for GFP fluorescence. Around the time of the meiosis II division, many cells (both wild type and mutant) contain four small GFP dots, most likely representing Spo14 near SPBs, as reported previously (Figure 10, A and D) (Rudge et al. 1998). (SPB staining was not observed in cells containing single-copy SPO14–GFP.) At later times during sporulation in wild type, circles of Spo14–GFP were observed; these eventually enlarged to fill the entire ascus (Figure 10, B and C). In ssp2 overproducing Spo14–GFP, a subset of cells contained large rings and were indistinguishable from the mature asci observed in wild type (Figure 10E). The frequency of cells that exhibit normal Spo14–GFP staining (∼23%) is similar to the frequency of cells that completed sporulation (∼21%). The remaining cells contained rings that were aberrant in several respects. First, the rings did not fill the entire ascus. Second, some rings had aberrant protrusions (Figure 10, F and G, arrowheads), reminiscent of the anucleate spore-like structures observed in the EM. Third, some of the cells were irregular in shape (Figure 10G, arrow), similar to the foot-shaped spores observed in the EM (Figure 8J). In every case, Spo14–GFP staining was more continuous in ssp2 cells containing multiple copies of the SPO14–GFP gene than in cells carrying an integrated version of SPO14–GFP.
Figure 10.—
Overproduction of Spo14 partially suppresses ssp2. Wild-type (A–C) and ssp2 (D–G) strains overexpressing SPO14–GFP were sporulated. They were then analyzed by observing GFP fluorescence in living cells. Bar, 2 μm. Open arrowheads in F and G point to aberrant protrusions also observed in the EM (Figure 8, H and I, arrows). Arrow in G points to a spore that is aberrant in shape, reminiscent of images seen in the EM (e.g., Figure 8J).
Overproduction of Sso1 partially suppresses the sporulation defect of ssp2:
To determine whether the Sso proteins can function as suppressors, multicopy plasmids carrying SSO1 or SSO2 (Ruohonen et al. 1997) were introduced into wild-type, ssp2, and osw1 strains. Overproduction of Sso1 increases sporulation in ssp2 (from 5 to 22%), but has no effect on sporulation efficiency in wild type or osw1. Overproduction of Sso2 had no effect on sporulation in wild type, ssp2, or osw1. The differential effect of SSO1 and SSO2 overproduction is consistent with the finding that Sso1, but not Sso2, functions in meiotic cells (Jantti et al. 2002).
DISCUSSION
SSP2 and OSW1—two genes involved in spore morphogenesis:
We have characterized extensively the phenotypes of the ssp2 and osw1 mutants. Our data demonstrate that the Ssp2 and Osw1 proteins are not required for early events in meiosis, including recombination, chromosome segregation, and nuclear division. These results are consistent with those of Sarkar et al. (2002), who examined nuclear division and recombination in ssp2. Our data indicate that Ssp2 and Osw1 are required for the normal formation of PSMs and mature SWs. This defect in spore morphogenesis is consistent with the timing of expression of the SSP2 and OSW1 genes (Chu et al. 1998; Nag and Axelrod 1998), which are expressed at the same time as genes involved in PSM formation.
Osw1 may play a regulatory role in spore morphogenesis:
A striking feature of the osw1 null mutant phenotype is the asynchrony of spore morphogenesis among different spores in the same ascus. Some nuclei are never encapsulated by PSMs even at very late times in sporulation medium, although well-defined PSMs encapsulate other nuclei in the same ascus. Consistent with the heterogeneity observed in the EM, fluorescence microscopy demonstrates variation in the number and morphology of Don1 rings. Some asci contain spores with SWs of different thicknesses, providing another indication of a lack of coordination among spores within an ascus. OSWs are rarely seen in osw1; the aberrant shapes of some prospores might result from lack of the OSW for stabilization. Coluccio et al. (2004) also observed an absence of OSWs in osw1; they did not report any other defects.
Several other mutants have been described that display heterogeneous defects in the development of PSMs and SWs (Friesen et al. 1994; Krisak et al. 1994; Wagner et al. 1997; Straight et al. 2000; Coluccio et al. 2004). These include mutations in four genes encoding protein kinases, SMK1, SPS1, CAK1 and MPS1, as well as mutations in the SPO75 gene, which encodes an integral membrane protein. The spectrum of phenotypes displayed by the smk1 null mutant is very similar to that of the osw1 mutant (Friesen et al. 1994; Krisak et al. 1994). It is generally assumed that the Smk1 kinase acts in a signaling pathway to promote and coordinate the different stages in spore wall morphogenesis (Wagner et al. 1999). The similarity in phenotypes between the osw1 and smk1 mutants suggests that Osw1 also performs a regulatory function.
The location of the Osw1 protein is compatible with its hypothesized function in the regulation of PSM and SW development. Osw1 is found at SPBs around the time that the meiotic plaque is active in initiating formation of the PSM. At subsequent stages, it surrounds developing prospores, consistent with it being a component of the PSM.
Ssp2 affects PSMs and SWs:
Our study reveals two aspects of the ssp2 mutant phenotype. First, there is a defect in PSM development. PSMs are often observed encapsulating materials both inside spores and in the spaces between spores. Furthermore, the PSM component, Spo14, fails to localize properly. Second, the ISW is only half the thickness of its wild-type counterpart. This aberration presumably explains why the developing spores are not refractile to light and therefore not observed in the light microscope. Unlike the osw1 mutant in which different spores develop to different extents, the ssp2 mutant phenotype is fairly homogeneous with almost all spores exhibiting the same phenotype.
There are two possible interpretations of the ssp2 mutant phenotype. First, the Ssp2 protein might act at two different steps in spore morphogenesis, first during PSM development and later during SW assembly. A more economical interpretation is that the primary defect in ssp2 is in PSM formation and the alteration in the SW is a secondary consequence of the defect in the PSM. SW materials might not be properly deposited between the two layers of the PSM when the PSM is aberrant. A role for Ssp2 in PSM development is consistent with the localization of Ssp2 to rings surrounding prospores even at very early stages in spore morphogenesis.
The ssp2 mutant phenotype has been characterized previously by two groups (Sarkar et al. 2002; Coluccio et al. 2004). Both groups reported that the PSM develops properly. However, the development of anucleate spore-like bodies is a subtle phenotype that might easily have escaped detection, and the localization of PSM components was not examined previously. Neither group reported any difference in the thickness of the ISW, although no measurements were presented. The most striking difference between our results and those published previously is that we do see an OSW in ssp2, whereas both Sarkar et al. (2002) and Coluccio et al. (2004) described the failure to develop OSWs as the sole defect in ssp2. The difference between our results and those published previously might be due to differences in yeast strain background and/or to differences in the methods used to prepare samples for examination in the EM. Our results indicate that Ssp2 acts at a much earlier stage than previously thought.
Ssp2 acts in the vesicle fusion pathway of PSM formation:
Formation of the PSM involves two distinct processes (Neiman 2005). The first process, membrane initiation, is a homotypic fusion event in which post-Golgi vesicles fuse with each other. This event occurs at the meiotic plaque of the SPB, and the plaque is essential for the coalescence of vesicles to initiate a PSM. The second process, membrane expansion, involves heterotypic fusion events in which vesicles fuse with the developing PSM until the nucleus is fully encapsulated.
The ssp2 mutant defects are partially suppressed by overproduction of either Spo14 or Sso1. Since these proteins are involved in vesicle fusion during PSM formation (Riedel et al. 2005; Nakanishi et al. 2006), it is likely that Ssp2 also plays a role in vesicle fusion. As noted in the Introduction, Spo14 in both initiation and expansion of the PSM (Riedel et al. 2005; Nakanishi et al. 2006). Similarly, Ssp2 could act at both steps. In support of a role in PSM initiation, the ssp2 mutant produces pieces of membrane that are not associated with SPBs, as if vesicle fusion events are not obligatorily coupled to the meiotic outer plaque in the absence of Ssp2.
Other observations suggest that Ssp2 plays a role in membrane expansion. Although fully developed PSMs are often observed in the EM in ssp2 cells, large circles of Spo14–GFP staining are never observed by fluorescence microscopy. This discrepancy suggests that Spo14–GFP localization is aberrant in PSMs, with Spo14 localizing in a punctate pattern instead of uniformly throughout the PSM. The aberrant localization of Spo14 in the PSMs that surround nuclei implies that PSM development is perturbed subsequent to the initiation stage. In addition, it is noteworthy that the PSMs that form in the ssp2 mutant do not always capture a nucleus (Figure 8, H and I), suggestive of a failure at the expansion stage (Neiman 1998).
Acknowledgments
We thank Aaron Neiman, JoAnne Engebrecht, and Sirkka Keranen for providing plasmids. Oligonucleotides were synthesized by the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. We are grateful to members of the Roeder lab for helpful discussions and comments on the manuscript. In addition, we thank Li-Lin Du, JoAnne Engebrecht, and Aaron Neiman for helpful discussions and sharing unpublished data and protocols. We are grateful to Barry Piekos for his able assistance with electron microscopy. This work was supported by grant GM28904 from the United States Public Health Service to G.S.R. and by the Howard Hughes Medical Institute.
References
- Agarwal, S., and G. S. Roeder, 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. [DOI] [PubMed] [Google Scholar]
- Bailis, J. M., and G. S. Roeder, 1998. Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev. 12: 3551–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, N., B. Grimwade, P. B. Ross-Macdonald, E.-Y. Choi, K. Finberg et al., 1994. Large-scale analysis of gene expression, protein localization and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105. [DOI] [PubMed] [Google Scholar]
- Christodoulidou, A., P. Briza, A. Ellinger and V. Bouriotis, 1999. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 460: 275–279. [DOI] [PubMed] [Google Scholar]
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein et al., 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–703. [DOI] [PubMed] [Google Scholar]
- Chua, P. R., and G. S. Roeder, 1998. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359. [DOI] [PubMed] [Google Scholar]
- Coluccio, A., E. Bogengruber, M. N. Conrad, M. E. Dresser, P. Briza et al., 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 3: 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L., and P. Novick, 2002. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi, A. H., and W. S. Saunders, 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, R. E., and M. S. Esposito, 1974. Genetic recombination and commitment to meiosis in Saccharomyces. Proc. Natl. Acad. Sci. USA 71: 3172–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen, H., R. Lunz, S. Doyle and J. Segall, 1994. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 8: 2162–2175. [DOI] [PubMed] [Google Scholar]
- Gerst, J. E., 1999. SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell. Mol. Life Sci. 55: 707–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, E.-J. E., and G. S. Roeder, 2002. A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev. 16: 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, R., and T. C. Sudhof, 1999. Membrane fusion and exocytosis. Annu. Rev. Biochem. 68: 863–911. [DOI] [PubMed] [Google Scholar]
- Jantti, J., M. K. Aalto, M. Oyen, L. Sundqvist, S. Keranen et al., 2002. Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J. Cell Sci. 115: 409–420. [DOI] [PubMed] [Google Scholar]
- Krisak, L., R. Strich, R. S. Winters, J. P. Hall, M. J. Mallory et al., 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8: 2151–2161. [DOI] [PubMed] [Google Scholar]
- Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko and M. Knop, 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20: 6946–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, D. K., and J. Axelrod, 1998. Identification of yeast meiosis-specific genes by differential display. Methods 16: 423–433. [DOI] [PubMed] [Google Scholar]
- Nakanishi, H., M. Morishita, C. L. Schwartz, A. Coluccio, J. Engebrecht et al., 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 119: 1406–1415. [DOI] [PubMed] [Google Scholar]
- Neiman, A. M., 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A. M., 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas, M. E., and A. M. Neiman, 2002. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics 160: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch, K. P., A. Toth, M. Galova, A. Schleiffer, G. Schaffner et al., 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11: 1001–1009. [DOI] [PubMed] [Google Scholar]
- Riedel, C. G., M. Mazza, P. Maier, R. Korner and M. Knop, 2005. Differential requirement for phospholipase D/Spo14 and its novel interactor Sma1 for regulation of exocytotic vesicle fusion in yeast meiosis. J. Biol. Chem. 280: 37846–37852. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., and G. S. Roeder, 1990. Meiosis in asynaptic yeast. Genetics 126: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, K., S. A. Rudge, M. A. Frohman, A. J. Morris and J. Engebrecht, 1995. Phospholipase D signaling is essential for meiosis. Proc. Natl. Acad. Sci. USA 92: 12151–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald, P., P. S. Coelho, T. Roemer, S. Agarwal, A. Kumar et al., 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413–418. [DOI] [PubMed] [Google Scholar]
- Rudge, S. A., A. J. Morris and J. Engebrecht, 1998. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruohonen, L., J. Toikkanen, V. Tieaho, M. Outola, H. Soderlund et al., 1997. Enhancement of protein secretion in Saccharomyces cerevisiae by overproduction of Sso protein, a late-acting component of the secretory machinery. Yeast 13: 337–351. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sarkar, P. K., M. A. Florczyk, K. A. McDonough and D. K. Nag, 2002. SSP2, a sporulation-specific gene necessary for outer spore wall assembly in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 267: 348–358. [DOI] [PubMed] [Google Scholar]
- Seifert, H. S., E. Y. Chen, M. So and F. Heffron, 1986. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 83: 735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shonn, M. A., R. McCarroll and A. W. Murray, 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289: 300–303. [DOI] [PubMed] [Google Scholar]
- Sikorski, R., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A. F., A. S. Belmont, C. C. Robinett and A. W. Murray, 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Straight, P. D., T. H. Giddings and M. Winey, 2000. Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell 11: 3525–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym, M., J. Engebrecht and G. S. Roeder, 1993. Zip1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- Tachikawa, H., A. Bloecher, K. Tatchell and A. M. Neiman, 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi, H., and G. S. Roeder, 2002. The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 22: 3078–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, M., P. Briza, M. Pierce and E. Winter, 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151: 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, M., M. Pierce and E. Winter, 1997. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16: 1305–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]