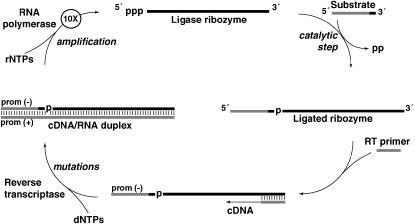
Figure 1.—
The continuous evolution (CE) scheme (Wright and Joyce 1997). RNA strands are solid and DNA strands are shaded. A population of ligase ribozymes (top) is incubated at 37° with an excess of substrate oligonucleotides in a 25-μl volume. Catalytically proficient ribozymes will ligate the substrate to their own 5′ ends (right). Also in the reaction vessel are a primer for reverse transcription that is complementary to the 3′ end of the ribozyme, a pool of dNTPs and rNTPs, and the protein enzymes MMLV reverse transcriptase and T7 RNA polymerase. While all RNAs should be reverse transcribed into double-stranded DNA/RNA hybrids (bottom and left), only those that had successfully performed ligation will be transcribed back into RNA because the substrate contains the necessary T7 promoter sequence. Because ∼10 RNAs are made from each template, and three completions of the cycle as shown occur in each 22-min burst, a 1000-fold amplification of RNA is possible if the initial population contains high-fitness genotypes. After 22 min, the reaction is diluted 1000-fold, fresh protein enzymes, primers, and nucleotides are added, and a new burst is initiated. The cDNA can be amplified via the PCR and genotyped each burst. If the mean fitness of the population falls, the degree of amplification will not keep pace with the dilution factor and eventually the cDNA concentration will drop below the PCR detection threshold.