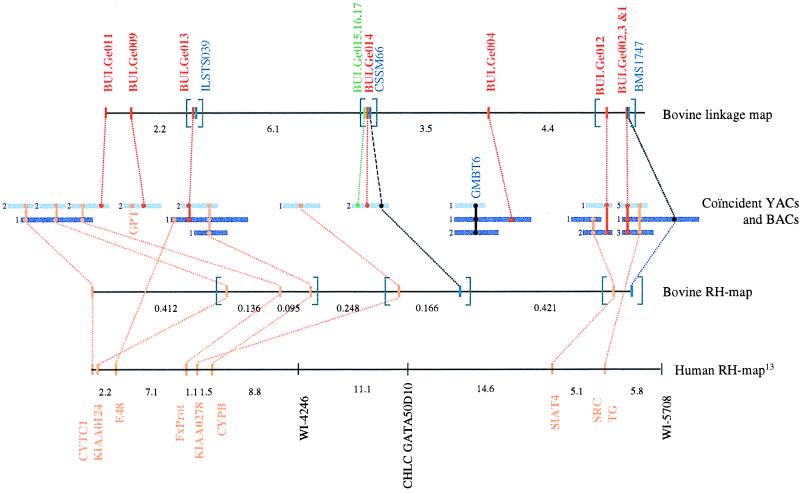
Figure 2.
Generation of a high-density marker map of BTA14q11–16. CATS were developed from genes positioned on the human RH map corresponding to HSA8q23-ter, shown in orange. The map position of the bovine orthologues was verified by using a hamster-bovine whole-genome RH panel. The corresponding bovine RH map is shown with most likely marker order and centirays between adjacent markers as estimated with rhmap. Marker sets that could not be ordered with odds >1,000 are in brackets. CATS mapping to BTA14q11–16 in cattle were used to screen a bovine YAC and BAC library. Resulting YACs are shown as dark blue bars, while BACs are shown as light blue bars. The numbers shown adjacent to the BAC and YAC clones correspond to the number of clones with identical sequence tagged site content. GMBT6, a variable number of tandem repeat known to map to BTA14q11–16 (29) also was used to screen the YAC and BAC libraries. Microsatellites and SNPs were isolated from the large insert clones as described and used to generate the illustrated linkage map. Most likely marker order and recombination rates between adjacent markers are shown. Marker sets that could not be ordered with odds >1,000 are in brackets. Newly developed microsatellites are shown in red, previously described markers in blue, and SNPs in green.