Figure 1.
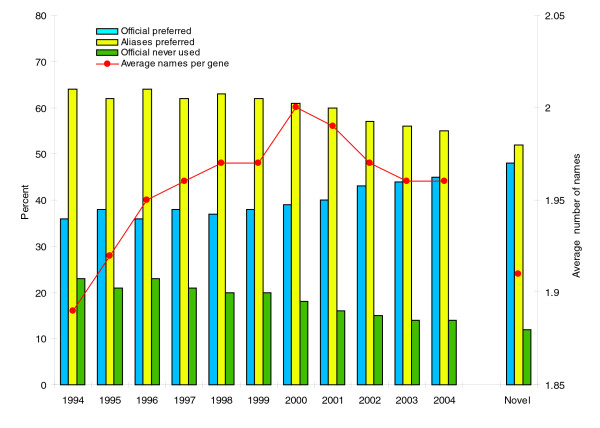
Usage of HUGO nomenclature in the past ten years. We analyzed PubMed abstracts for the period 1994-2004, collecting information about the human genes mentioned on the abstracts, and noting how such mention was made (official symbol or other aliases). Names were detected using Text Detective (BioAlma SL), a gene name recognition software that is able to recognize human gene names in texts with high recall and precision, distinguishing real instances of the gene from other uses and meanings of the same name [13]. Text Detective combines gene name recognition with standardization of citations, using HUGO nomenclature in the case of human genes. Additional results (the yeast results discussed in the text) were obtained using the Information Interlinked Over Proteins (iHOP) system [14] in order to discard possible biases due to the name-recognition software used. The percentage of genes that are cited predominantly by their official name is used as a measure of the support for official names. Blue bars show the percentage of genes for which the official name is favored (the official name is mentioned more often than aliases). Yellow bars show the inverse, the percentage of genes for which aliases are favored. Green bars show the percentage of cases in which the official name is never used, and all mentions correspond to aliases. Also, the average number of names per gene is shown, computed as the total number of names used divided by the total number of genes. The last column, labeled 'Novel', takes into account only those genes whose first mention in the literature occurred in the year 2000 or later.
