Figure 1.
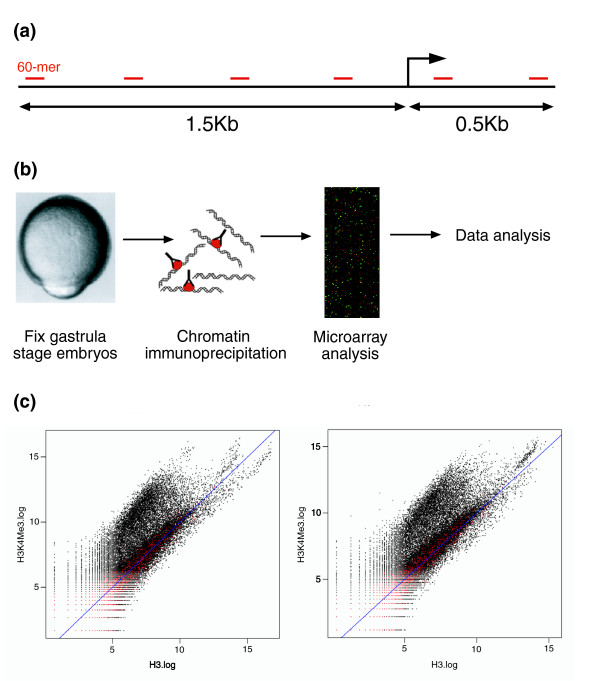
ChIP-chip method in zebrafish embryos. (a) Design of promoter arrays: 60-mer oligonucleotides were designed against genomic sequence 1.5 kb upstream and 0.5 kb downstream of the annotated transcription start site of approximately 11,000 zebrafish genes. The resulting probes are arrayed onto two microarray slides. (b) ChIP-array protocol. (c) Examples of scatter plots obtained from one hybridization of immunoprecipitated DNA on one 2-slide proximal promoter microarray set. Log2 ratios for each labeled sample are plotted against each other. Enriched probes are seen above the diagonal. Control spots (zebrafish gene desert and Arabidopsis gene probes), shown in red, fall along the diagonal.
