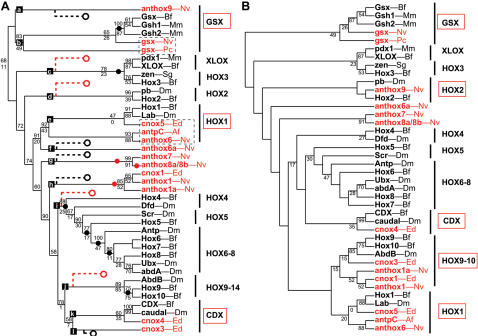
Figure 9.
Re-analysis of the Kamm et al. maximum-likelihood phylogeny. (A) The redrawn maximum-likelihood phylogeny presented by Kamm et al. [49]. The tree is unrooted. Based on the Dayhoff-PAM1 substitution matrix [129] the tree's overall likelihood is −2288.08809. Bootstrap proportions determined in the original study are shown above the relevant nodes. Bootstrap proportions determined in the current study are shown below the relevant nodes. Inferred lineage specific gene losses are represented by open circles (black for Bilateria and red for Cnidaria). Inferred lineage specific gene duplications are represented by solid circles (black for Bilateria and red for Cnidaria). Sequences inferred to have been present in the common ancestor are indicated by lettered squares. The names of gene families that contain cnidarian representatives are enclosed by red lines. (B) A tree we identified using global instead of local rearrangements with the Kamm et al. data and the Dayhoff substitution matrix. The tree's overall likelihood is −2280.76406. Bootstrap proportions determined in the present study are shown below the relevant nodes. The dataset used in this analysis is available as Figure S13.