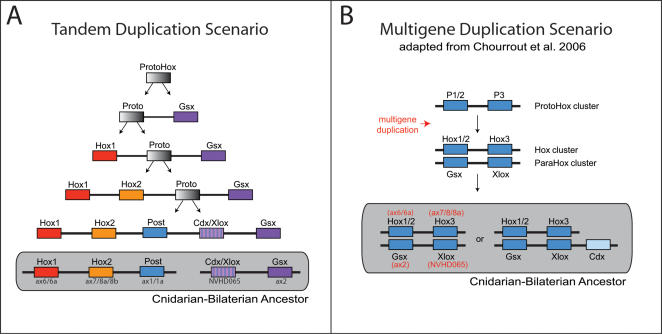
Figure 11.
Origins of the Hox and ParaHox clusters. (A) The tandem duplication model of Hox and ParaHox clusters. Our evidence conflicts with the current theory that a multigene duplication event formed the Hox and ParaHox cluster. Under this alternative model, each Hox and ParaHox gene was formed through a series of tandem duplications either in the 3′ or 5′ direction and a translocation event separated the two ParaHox genes from the Hox cluster. Hox3 and Central Hox genes are not included in this model because it is not clear whether they were present in the cnidarian-bilaterian ancestor and subsequently lost in the Cnidaria or alternatively, were derived in the Bilateria from subsequent tandem duplication events of another Hox/ParaHox gene (for example anthox7/8a/8b or anthox1/1a). The general scenario presented here is robust enough to easily accommodate either event. (B) In this scenario adapted from Chourrout et al. 2006 Figure 3, a two-gene ProtoHox cluster is duplicated to form two sister clusters, the Hox and ParaHox clusters [51]. Chourrout et al. found that NVHD065 (Xlox/Cdx in their analysis) showed conflicting affinities for Xlox and Cdx; similarly our neighbor-joining and Bayesian trees had NVHD065 grouping with Cdx while the maximum-likelihood tree had NVHD065 grouping with Xlox. Unlike our study, Chourrout et al. considered anthox1 and anthox1a (HoxE and HoxF in their analysis) to be non-Hox/ParaHox lineages, hence their omission from their model [51].