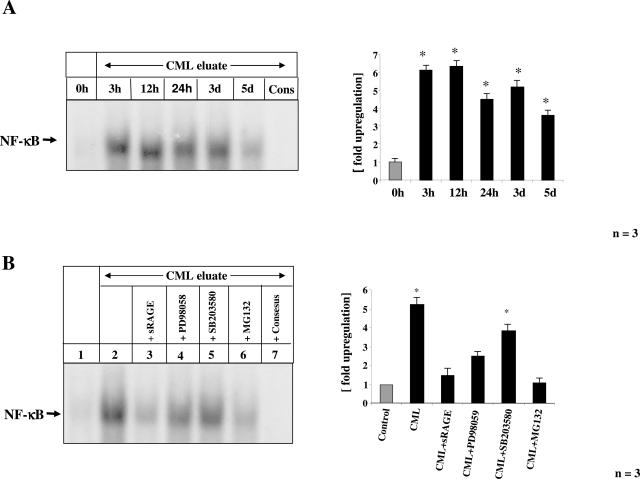
FIGURE 6.
Time course of CML-mps induced activation of NF-κB in cultured endothelial cells. Cultured BAECs were incubated with CML-mps (100 μg) eluted from highly inflamed areas from CD patients for the time points indicated in the figure legend before cells were harvested and nuclear extracts were prepared. NF-κB binding activity was determined by EMSA. Specificity of NF-κB binding (using the same sample as in lane 5 in A) was shown by including a 160-fold molar excess of unlabeled consensus NF-κB oligonucleotide (Cons). The experiment was repeated three times with identical results and the results of one representative patient are shown on the left. The bar graphs on the right summarize the results and densitometric analysis of all extracts tested. The mean ± SD is reported. B: Cultured BAECs were preincubated with either sRAGE (25 μg), the p44/p42 MAPKinase inhibitor PD98058 (30 μmol/L), the p38 MAPKinase inhibitor SB203580 (10 nmol/L), and the proteasome inhibitor MG132 (1 μmol/L) 2 hours before stimulation with CML-mps eluates isolated from highly inflamed areas from CD patients for 3 days. NF-κB binding activity was studied in EMSA. The experiment was repeated three times with identical results, and the results of one representative patient are shown on the left. Specificity of NF-κB binding (using the same sample as in lane 2) was shown by including a 160-fold molar excess of unlabeled consensus NF-κB oligonucleotide (lane 7). The bar graphs on the right summarize the results and densitometric analysis of all extracts tested. The mean ± SD is reported.