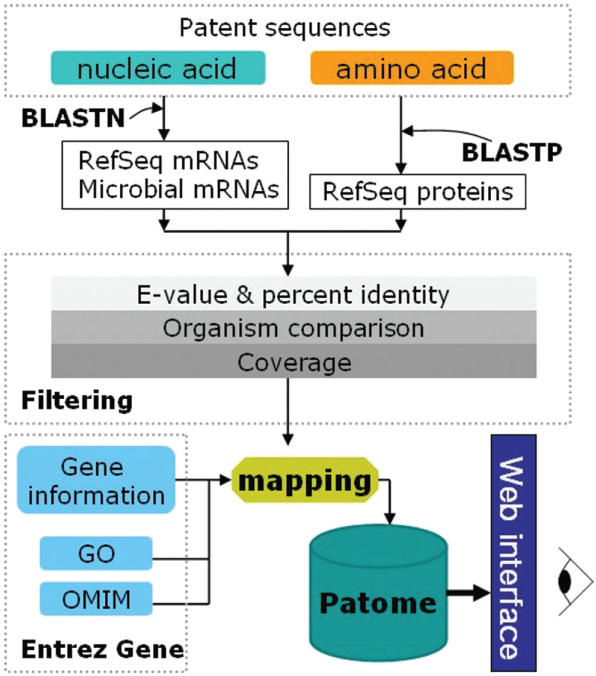
Figure 1.
Overview of the analysis flow in Patome. The downloaded sequences are divided into nucleic acids and amino acids. Nucleic acids are annotated with both RefSeq mRNAs and microbial mRNAs. Amino acids are annotated with RefSeq proteins. The BLAST results are filtered with three filtering methods: E-value & percent identity, organism comparison and coverage. From the mapping between the annotated data and biological database information (Gene, GO, OMIM), a gene–patent association table is built. The Patome database is constructed to store annotation and analysis data and to make this information available to users via web interfaces.