Figure 3.
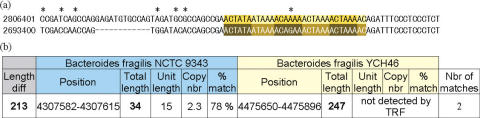
Examples of uncertain locus length. (a) Visualisation of the alignment of a possible VNTR loci. The upper sequence is from Bacteroides fragilis strain NCTC 9343, and the lower sequence is from B.fragilis strain YCH46. A 12 bp gap can be observed in this alignment. Because only a few of nucleotides exist between the gaps and the genuine TR, the primers must be designed to the 5′ of these gaps. Although the aligned repeats of both loci, which we call alignment length, are both exactly 31 bp, the PCR products will have a 12 bp difference. In VNTRDB, these 12 bp difference will affect the raw length but not the reported alignment length. (b) The loci were obtained directly from GPMS. Although total lengths of loci are different between these two Bacteroides strains, users cannot be sure whether this length difference is due to copy number difference alone or other sequence insertions or deletions. In VNTRDB, the corresponding locus in YCH46 has a locus length of 28 bp and raw length 244 bp suggesting that there are insertion/deletions resulting from changes other than repeat copy number differences. This combined with the fact that the visualization tool can only produce an alignment with very permissive parameters informs the user that the similarity of flanking sequences is very low and therefore the locus is probably not a genuine VNTR.