Abstract
Annotation of protein-coding genes is a key goal of genome sequencing projects. In spite of tremendous recent advances in computational gene finding, comprehensive annotation remains a challenge. Peptide mass spectrometry is a powerful tool for researching the dynamic proteome and suggests an attractive approach to discover and validate protein-coding genes. We present algorithms to construct and efficiently search spectra against a genomic database, with no prior knowledge of encoded proteins. By searching a corpus of 18.5 million tandem mass spectra (MS/MS) from human proteomic samples, we validate 39,000 exons and 11,000 introns at the level of translation. We present translation-level evidence for novel or extended exons in 16 genes, confirm translation of 224 hypothetical proteins, and discover or confirm over 40 alternative splicing events. Polymorphisms are efficiently encoded in our database, allowing us to observe variant alleles for 308 coding SNPs. Finally, we demonstrate the use of mass spectrometry to improve automated gene prediction, adding 800 correct exons to our predictions using a simple rescoring strategy. Our results demonstrate that proteomic profiling should play a role in any genome sequencing project.
Annotation of protein-coding genes is a key goal of genome sequencing projects. In spite of recent advances in computational gene finding, a comprehensive annotation of protein coding genes remains challenging. In most annotation pipelines, a computationally predicted gene must be confirmed by independent evidence and/or manual validation before it is accepted. The additional evidence is often in the form of conservation across distant organisms or evidence of transcription. This evidence, while compelling, is not sufficient (see Gupta et al. 2004). Conservation across species is not limited to protein coding regions. Roughly 5%–20% of the human genome is conserved against mouse, of which just 1%–2% is considered to be coding for proteins (Waterston et al. 2002). Likewise, most cDNA sequences are obtained from single-pass, high-throughput sequencing and contain sequencing errors, prespliced mRNA, as well as untranslated regions. Thus it is hard to determine if every alternative splice form predicted from an EST is also expressed at the protein level. Alternative splicing and overlapping genes present particularly difficult annotation problems. Some estimates suggest that the majority of human genes undergo alternative splicing (Mironov et al. 1999; Modrek and Lee 2002; Florea et al. 2005).
Therefore, it is customary to provide a conservative genome annotation and then rely upon community efforts to refine annotations and fill in missing genes. While the genome annotation process is unlikely to be fully automated, high-throughput methods are an important part of any genome annotation strategy. Tandem mass spectrometry is an attractive technique for validating gene predictions. It measures proteins directly, verifying putative gene products at the level of translation. Also, it provides an orthogonal line of evidence, with different error sources than nucleotide-based approaches.
A tandem mass spectrum can be viewed as a collection of fragment masses from a single peptide (eight to 30 amino acids from an enzymatically digested protein). This set of mass values is a “fingerprint” that identifies the peptide. The spectra are usually not analyzed de novo. Instead, they are compared against peptides from a database of known proteins (Aebersold and Mann 2003). Much research has been devoted to improving the accuracy of this search by refining scoring (Yates et al. 1995b; Perkins et al. 1999; Bafna and Edwards 2001, Creasy and Cottrell 2002; Lu and Chen 2003; Sadygov and Yates 2003; Tabb et al. 2003), improving search speed (Craig and Beavis 2003; Frank et al. 2005), and handling post-translational modifications (Tsur et al. 2005).
In this context, it is natural to ask if we can search translated genomic databases directly. Each match from such a search confirms a genomic locus to be part of a protein-coding gene. This has been proposed in a number of studies (Yates et al. 1995a; Choudhary et al. 2001; Kuster et al. 2001; Carlton et al. 2002; Fermin et al. 2006). However, in eukaryotes, searching a straightforward six-frame translation is problematic. The typical exon in a multi-exonic gene is short, with an average length of 150 bp (50 amino acids). A significant fraction (∼25%) of trypsin-digested peptides from eukaryotes span an exon boundary and so cannot be identified with an ORF database. Predicting the correct introns is a difficult step in gene finding, and such exon-spanning peptides are critical to confirming and annotating splicing. Also, only a small fraction of the genome codes for proteins. A six-frame translation of the human genome has 6-Gb residues, while the size of the known human proteome is just 25 Mb. Scaling up to a larger database makes searches slower by orders of magnitude. In addition to the issue of speed, searches are known to have a significant error rate, and larger databases incur a higher false-positive rate. Polymorphisms are also a potential source of error in such a search.
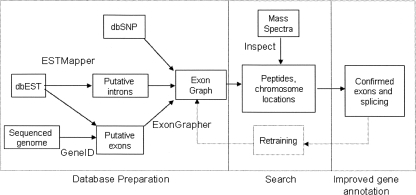
We overcome these issues with several technical improvements. First, instead of searching translated genomes directly, we search a compact representation of all putative exons, splice variants and polymorphisms. This representation takes the form of a directed acyclic graph which we call the exon graph. Our search is efficient, using a database filtering technique based on tagging (Frank et al. 2005) that extends directly to searching graphs instead of sequences. We also use improved scoring (Keller et al. 2002; Tanner et al. 2005) to keep the false discovery rate at 2.5%. We show that evidence from mass spectrometry can be fed into computational gene finding methods to improve gene predictions. An outline of our method is presented in Figure 1.
Figure 1.
Overview of the workflow for genome annotation through mass spectrometry. The exon graph database is constructed without reference to prior annotations of the genome. Putative exons and exon-pairs are generated through EST alignment and de novo predictions; homology maps are another potential source. Peptide matches identify the true exons (and introns) among the gene predictions
Methods
Exon and intron predictions
Exon predictions were generated by GeneID (Parra et al. 2000; Blanco et al. 2002) against build 35 (May 2004) of the human genome. All putative exons with a score of ≥−1 were retained, producing 4,110,476 exons with considerable overlap. Splice junctions were considered between all pairs of exons with compatible reading frames and intron length between 25 and 20,000 bases. Each interval was linked to the closest intervals with a compatible reading frame. At most 10 introns were considered per genomic position.
We extracted human sequences from dbEST (6,587,476 sequences) (Boguski 1993). These sequences were aligned against the May 2004 assembly of the human genomic sequence using ESTMapper (Florea et al. 2005). A total of 7,153,771 alignments were generated (including multiple alignments for some sequences). Because genomic contamination and sequencing errors produce noise in EST data, we filtered the set of putative exons and introns. Mappings with sequence identity <90% or containing cDNA gaps were removed. We also compared each splice junction against the consensus splice signal using a position weight matrix. We discarded any putative intron that (1) occurred in only one EST mapping and (2) had a poor (fifth percentile or less) signal score. Roughly 10% of all introns (336,833) were discarded in this way. The signal score and occurrence count of each intron are stored in the database for later reference. After filtering, 6,923,229 EST mappings were generated, with an average of 2.2 intervals per EST.
Database construction
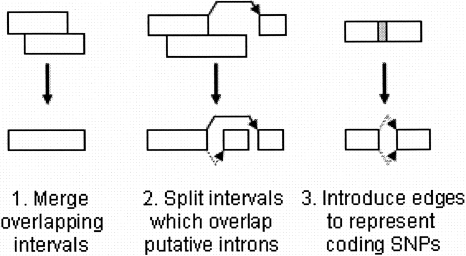
Our goal is to build a compact representation of all the exons and introns derived from GeneID and ESTMapper. Let I and J be the collection of intervals and splice junctions for a chromosome strand. The endpoints of interval In are denoted as Ln and Rn, with the convention that In includes the bases from Ln up to (but not including) Rn. We call a point a “junction point” if it is an edge of any putative intron. Refer to Figure 2 for an overview of the procedure, and Figure 3 for an example of the final graph.
Figure 2.
Overview of the procedure for turning a collection of putative exons and introns into an exon graph. Adjacent edges are represented by dotted lines, splice events by solid lines.
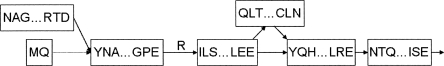
Figure 3.
A portion of the exon graph for heterogeneous nuclear ribonuclear protein K. The labeled edge represents a codon split across a splice junction. The dotted edge is an “adjacent edge” corresponding to a longer form of an exon. Searching the exon graph reveals peptides spanning both outgoing edges from the central node, confirming alternative splicing at the level of translation.
Gene prediction algorithms often produce putative exons of various lengths which overlap. Similarly, because ESTs have varying read lengths, it is common for them to map to overlapping genomic intervals. If intervals Ii and Ij overlap, we can merge them into a larger interval without loss of information, so long as
Li = Lj, or max(Li,Lj) is not a junction point.
Ri = Rj, or min(Ri,Rj) is not a junction point.
We perform all such legal merges. This phase greatly reduces the redundancy of the set of intervals. If an interval overlaps the edge of a putative intron, we cut the interval into two subintervals at the junction point. At the end of this phase, our set of intervals is disjoint. We now add an edge between any adjacent intervals (Ii and Ij such that Ri = Lj). For each putative intron, we add a splice edge between the corresponding intervals. We now incorporate polymorphisms. If an interval contains a coding SNP, we add intervals for each allele. Thus, each SNP produces a “bulge” in the graph.
We derive an exon graph from the genomic interval graph. For each node in the interval graph, add one node to the exon graph for each legal reading frame. Each exon graph node has a protein sequence and may have an untranslated prefix and suffix. If intervals are joined by an edge, then the corresponding exons (with compatible reading frame) are similarly joined. Edges are annotated with an amino acid when a codon is split between exons.
In order to remove noncoding “noise” from the database, we remove all nodes and edges that are not part of a coding sequence of length 50 or more. This procedure removes nodes corresponding to translation of EST mappings in the wrong reading frame. The finished exon graph contains a total of 133 M amino acids, in 3.5 M exons, with 2 M splice junctions.
Mass spectra
Proteins were extracted from HEK293 cell culture. Our standard extraction contains 2% RapiGest (Waters) in TNE buffer. Disulfide bonds were reduced using a final concentration of 2 mM TCEP for 30 min. A final concentration of 5 mM iodoacetamide was used to alkylate sulfhydryl groups. Protein concentration was measured with a Bradford assay. Proteins were then digested with trypsin (1:50) overnight.
An Agilent 1100 HPLC system (Agilent Technologies) was used to deliver a flow rate of 300 nL min−1 to the mass spectrometer through a splitter. Chromatographic separation was accomplished using a three-phase capillary column. Using an in-house constructed pressure cell, 5 μm Zorbax SB-C18 (Agilent) packing material was packed into a fused silica capillary tubing (200-μm inner diameter (ID), 360-μm outer diameter (OD), 20 cm long) to form the first dimension RP column (RP1). A similar column (200-μm ID, 5 cm long) packed with 5 μm PolySulfoethyl (PolyLC) packing material was used as the SCX column. A zero dead volume 1-μm filter (Upchurch, M548) was attached to the exit of each column for column packing and connecting. A fused silica capillary (100-μm ID, 360-μm OD, 20 cm long) packed with 5 m Zorbax SB-C18 (Agilent) packing material was used as the analytical column (RP2). One end of the fused silica tubing was pulled to a sharp tip with the ID <1 μm using a laser puller (Sutter P-2000) as the electro-spray tip. The peptide mixtures were loaded onto the RP1 column using the same in-house pressure cell. To avoid sample carryover and keep good reproducibility, a new set of three columns with the same length was used for each sample. Peptides were first eluted from the RP1 column to the SCX column using a 0%–80% acetonitrile gradient for 150 min. The peptides were fractionated by the SCX column using a series of salt gradients (from 10 mM–1 M ammonium acetate for 20 min), followed by high-resolution reverse phase separation using an acetonitrile gradient of 0%–80% for 120 min. We have found that a three-dimensional run can provide significantly more resolving power but at the cost of a longer separation time. For three dimensions, we elute fractions with acetonitrile from RP1 in 10% increments and then perform the salt elutions as described above but with a resolving gradient for RP2 of acetonitrile equal to the gradient used to elute from RP1.
Spectra were acquired on LTQ linear ion trap tandem mass spectrometers (Thermo Electron Corporation) employing automated, data-dependent acquisition. The mass spectrometer was operated in positive ion mode with a source temperature of 150°C. As a final purification step, gas phase separation in the ion trap was employed to separate the peptides into three mass classes prior to scanning; the full MS scan range was divided into three smaller scan ranges (300–800, 800–1100, and 1100–2000 Da) to improve dynamic range. Each mass spectrometry (MS) scan was followed by 4 MS/MS scans of the most intense ions from the parent MS scan. A dynamic exclusion of 1 min was used to improve the duty cycle.
In addition, we downloaded all human, non-ICAT-labeled spectra publicly available (as of March 2006) in the PeptideAtlas data repository (Desiere et al. 2004). These data consist of spectra from the erythroleukemia K526 cell line (Resing et al. 2004), and from the HUPO Plasma Proteome Project (Omenn et al. 2005). The data include a total of 1.8 million spectra in 621 MS runs, most of them from ion trap mass spectrometers.
The HEK293 mass spectra are available from http://bioinfo2.ucsd.edu, together with spectrum annotations.
Database search
The database search proceeds by a modified version of the Inspect search algorithm (Tanner et al. 2005). Given a spectrum, we perform partial de novo reconstruction to generate a peptide sequence tag of three or more amino acids. To accommodate de novo errors, we generate multiple tags and store them in a trie (Aho and Corasick 1975). When a tag sequence is found in the database, we perform a depth-first search in the graph to find all extensions that match the tag’s flanking masses. The source code for our software is available from our laboratory’s Web page (http://peptide.ucsd.edu/).
When a tag and its flanking masses are matched, a candidate peptide is produced. Each candidate peptide is scored to compute the probability of that peptide generating the query spectrum (Tanner et al. 2005). Inspect computes match quality scores based upon fragment presence and intensity, and the presence of unexplained “noise” peaks. Once the database scan is complete, the top matches are reported. If the same peptide sequence is observed multiple times, up to 10 loci matching the peptide are reported. To improve filtering of incorrect matches, we also consider the difference between the top match score and the score of the next-best peptide (delta-score). To correct for the dependence of delta-scores on database size, we take the ratio of a match’s delta-score to the average delta-score across all matches. The weighted sum of the match quality score and delta-score is called an F-score.
The empirical distribution of F-scores can be fit by a mixture model of a gamma distribution (representing false annotations) and a normal distribution (representing true annotations) (Keller et al. 2002). We select an F-score cutoff which corresponds to a P-value of 0.05 (95% probability of correct annotation).
As an additional measurement of false discovery rate, we constructed a reversed database by reversing the sequences of all nodes and reversing the direction of each edge. We measured an empirical false discovery rate by searching 700,000 spectra against the reversed databases. Our F-score cutoff yields 1200 matches on the reversed database, for a false annotation rate of 0.2%. In a search of the forward database, 47,000 spectra passed this same score cutoff. Based on these results, we estimate that 1200 of the 47,000 spectrum matches against the true database are incorrect, for a false discovery rate of 2.5%. In addition to this filter at the spectrum level, we pay particular attention to exons hit by multiple peptides; no such instances were observed for the search of the reversed database.
Post-processing of the search results was performed to deal with peptides which occur in multiple proteins. We note that in addition to closely related paralogs, the predicted exons may include some pseudogenes highly similar to their source genes. As an extreme example, the peptide AMGIMNSFVNDIFER (from H2B histone family, member S) is found in >20 valid and invalid ORFs. Therefore, when measuring coverage, we iteratively select a set of genes. At each stage, the gene which can be used to annotate the greatest number of spectra is selected, and the selected gene “absorbs” all shared peptides. We require at least two peptide hits before judging a protein present. This procedure ensures that redundant or questionable protein records are not selected. When considering alternative splicing, we select multiple isoforms of a protein only if we must do so in order to account for all the peptides matched.
Mapping known proteins to the genome
We wish to ensure the exon graph database captures the exons and introns from known genes. To do this, we produce the full genomic alignment of each protein, including splice junctions. We first identify “seeds,” positions on the genome which appear to match the protein. The chromosome locations are stored for most (54,032) of the IPI database records. In addition, each protein was searched against the repeat-masked human genome using TBLASTN. Finally, the exon graph was searched for any gene containing length-8 substrings (words) from the full protein; the three records with best coverage were retained as seeds. As a filtering step, we consider only seed matches that cover at least 30 residues of (an exon from) the source protein.
The heuristic alignment algorithm enumerates 6-mers from the protein found in the six-frame translation of the genomic region of interest. Adjacent hits are merged into putative exons. Using dynamic programming, we find a chain of exons which cover the entire protein. Exons close to each other can be merged, to step over mismatches between the protein sequence and genome. Finally, exon endpoints are refined to capture the best available splice signals.
A total of 56,725 proteins (98%) were mapped against the genome with ≥95% sequence identity. Of these, 37,849 (65%) were mapped with 100% identity. Of the records not successfully aligned, many have no satisfactory “seed” in the TBLASTN results. Records that represent short signal peptides are often missed in this way (data not shown). Many of the nonaligned proteins are predicted protein sequences derived from cDNA, which may be chimeric.
Each peptide identified in our database was compared to the locations of known proteins. If a peptide was found multiple times in the genome, or if two matches had equivalent match scores, we considered each locus. When selecting a locus, the order of preference was as follows: match to a known gene, match a known gene with SNPs, match a novel single-exon peptide, match a novel intron-spanning peptide. This procedure helps us avoid proposing new exons which correspond to pseudogenes.
Improving gene predictions
Our goal was to demonstrate automated refinement of gene prediction by incorporating MS search results. We selected the GeneID software because it uses a simple two-pass approach to gene prediction. It first predicts a collection of coding exons and then chains these exons into complete genes. Our strategy is to search the exon graph and then boost the scores of exons and introns that correspond to peptides. The assignment of peptide matches to known genes was not used when improving gene predictions.
We first ran GeneID against the human genome, retaining all predicted exons with score ≥−5. We note that exon scores are derived from a log odds ratio; GeneID attempts to avoid incorporating exons with negative scores. We then examined the number of peptide matches which hit each exon, and the P-value of these matches. We note that if the coding sequence for a peptide spans exons, one of which accounts for just one base pair, there may be several plausible exon pairings that encode the same peptide. Therefore, to reduce false positives, we register an exon hit only if the peptide match is “anchored” by at least 7 bp on the exon.
For each exon, we consider three parameters. The parameter c is equal to the number of spectrum annotations that are contained in the exon of interest. The parameter Pa is set to the best P-value of a peptide match covering the splice acceptor of the exon. We set Pa = 1 if there are not at least two spectrum annotations covering the acceptor site. Otherwise, we add 0.001 to the P-value to limit the effects of matches with extremely low P-values. Similarly, Pd is the best P-value of a match covering the splice donor. The score S of each exon is modified as follows:
The weights w1 and w2 were tuned to 1.0 and 0.8, respectively, by computing accuracy over a test set of 100 genes from chromosome 1.
For each gene of interest, we extract the genomic interval containing the exons from the gene. We run GeneID in exon-chaining mode to predict a gene on this interval using the original exons, then using the rescored exons.
Results
Search algorithm comparison
We compared the performance of Inspect to that of SpectrumMill (version 3.1, Agilent) on a collection of 800,000 spectra (34 runs) from the HEK293 data set. Both tools searched these spectra against the same database consisting of the IPI database, together with the reversed sequence of each protein. We assume that spurious matches are distributed randomly throughout the database. Using this assumption, if 5% of all matches come from reversed proteins, then the false discovery rate among matches from valid proteins is also 5%. Sorting the SpectrumMill matches by score, we obtain 94,633 spectrum annotations (27,845 distinct peptides) at a false discovery rate of 5%.
Sorting the Inspect matches by score, we obtain 135,192 spectrum annotations (43,311 distinct peptides) at this same false discovery rate. These results (40% more spectra, 70% more peptides) indicate that Inspect’s filtering and scoring are effective on this data set.
Exon graph construction
One goal in building the exon graph database is to keep the database size as small as possible while still covering all splice variants of all genes. The exon graph contains a total of 134 million amino acid residues, a significant savings over the full length of the EST database (2 billion residues), or the concatenated exon predictions from GeneID (630 million residues). The graph contains a total of ∼3 M exon nodes and ∼8 M edges. Modeling possible splicing events as a graph is a familiar formalism (Heber et al. 2002; Leipzig et al. 2004), although our construction of the exon graph differs from previous work (see Methods).
To verify the completeness of the exon graph, we considered the IPI database (version 3.15) as a representative corpus of known human proteins (Kersey et al. 2004). The IPI database contains 25 million residues in 58,099 records. We note that the database is not complete and contains some hypothetical sequences. We aligned these proteins against the human genome using known genomic locations and BLAST, as described in the Methods. We restrict our attention to the 56,725 records mapped to the human genome at 95% or greater sequence identity, the “mapped proteins.” We use this large reference set to estimate the proportion of known genes contained in the exon graph.
The mapped proteins include multiple isoforms of many genes. Counting known proteins that share exons as one gene, we reach a gene count of 32,493, of which 10,583 have multiple isoforms (Supplemental Fig. 1). These gene mappings include a total of 442,572 distinct exons. We show later the annotation of peptides corresponding to isoforms that are not contained in the IPI database but have been deposited in GenBank.
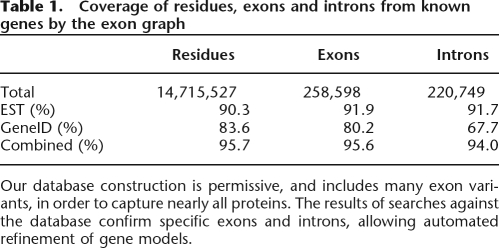
For each mapped protein, we determined whether GeneID predictions and/or EST mappings captured the genomic intervals (exons) and putative splice junctions (introns) of the protein. Table 1 summarizes the results.
Table 1.
Coverage of residues, exons and introns from known genes by the exon graph
Our database construction is permissive, and includes many exon variants, in order to capture nearly all proteins. The results of searches against the database confirm specific exons and introns, allowing automated refinement of gene models.
This table reflects the extremely high EST coverage of the human proteome. The exon predictions from GeneID cover most true exons, but the intron coverage is lower. The low intron coverage likely results from the simplistic exon-joining algorithm used in constructing the exon graph. A more sophisticated approach may cover more splice junctions. The exons missed in this construction typically come from the edges of the protein. The coverage rates for first and last exons are 81% for ESTs and 60% for GeneID, significantly lower than the average overall. Further research will target these problematic exons. Given the high coverage of known proteins by the algorithmically derived exon graph, we turn now to the results of mass spectrometric annotation with the exon graph.
Search results
We obtained ∼18.5 M spectra from various tissue types and searched them against the exon graph. Searches of this large data set were run over a grid of 1.6-GHz compute nodes (FWGrid Project). The average search time on a node was ∼2.5 sec per spectrum. Low-quality matches were filtered out a threshold based on the distribution of match scores (see Methods). Matches shorter than eight amino acids were discarded due to the difficulty in assigning short peptides to a unique locus.
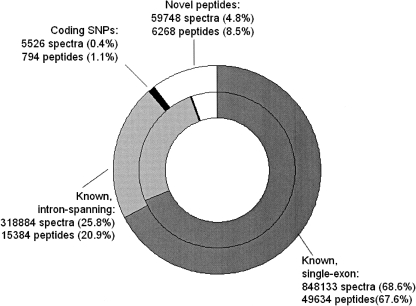
Each annotation includes the genomic location of the peptide. We compare these loci to the chromosomal locations of known proteins. We then categorize peptide matches based upon their relationship to known genes (see Methods). Recall that the human genome is heavily annotated. Therefore, the degree to which known proteins are covered by annotations from this data set is a reasonable estimate of our coverage of the full proteome. See Figure 4 for an initial breakdown of the results. The majority (89%) of peptides match known genes. Of these, 24% span an exon boundary, confirming splicing events at the protein level. A total of 121 peptides (in 1517 spectra) span two exon boundaries; these represent cases where a tryptic peptide fully spans a short exon. A total of 11,050 splice events are confirmed by identified peptides. Given that only ∼20% of the exon graph corresponds to known proteins, the enrichment for known genes suggests that protein-coding regions of unannotated genomes can be discovered by these methods. Those peptides that do not match known genes may be discoveries of novel exons, or novel splicing events; these cases are discussed after the results from known genes.
Figure 4.
Categorization of search results by their relationship to known proteins. The inner ring shows findings at the spectrum level; the outer ring shows findings at the level of distinct peptides. The peptides categorized as “unknown” include some peptides missing from the IPI database, as well as novel exons.
Protein coverage
The search results include 6252 proteins confirmed by two or more distinct peptides, and a total of 3745 proteins are matched by five or more distinct peptides. As noted earlier, we select a minimal set of proteins which account for spectrum annotations. This allows us to avoid listing records corresponding to multiple isoforms of the same protein unless both forms are in fact present.
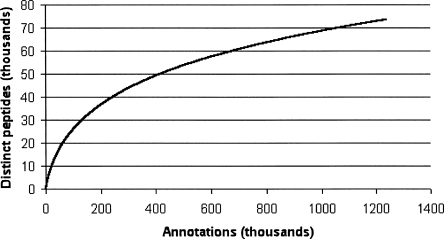
Because protein abundances within the cell vary greatly, we see extreme variation in the number of spectra matching each protein, with >25,000 matches from enolase 1, (alpha) but only one or two matches to other proteins. As with other high-throughput techniques such as cDNA sequencing, the repeated sampling of common elements eventually reaches saturation. We count the number of distinct peptides (from known proteins) discovered for a given number of identifications and plot the resulting discovery curve. The discovery rate slows as more peptides are found (Fig. 5), but is still far from saturation. The discovery curve is fit well by the function y ∼ x0.55 (correlation coefficient 0.97). Sampling of proteins from more tissue types promises to yield annotations for a wider range of proteins. Based on this discovery curve, we estimate that a 10-fold larger data set should yield high-confidence identifications (five or more distinct peptides) for >12,000 gene products.
Figure 5.
Discovery curve, plotting the number of distinct peptides as a function of the number of search hits.
Novel peptides
Matches to the exon graph which do not correspond to known proteins are potentially of great interest, since they may come from uncharacterized exons or even unannotated genes. We investigated and categorized all peptide matches that are not present in the IPI reference database. We reiterate that searching a larger database increases the likelihood of obtaining a high-scoring match by chance, and we employ several safeguards to filter such matches. First, we use a cutoff based on the false discovery rate (see Methods) to limit the number of such matches. Second, we used the results of a standard database search to filter any novel matches that can be explained away by a known peptide that is missing from the exon graph. An example of a peptide removed by this filtering is LGEHNVEVLEGNEQFINAAK, coded by an intron of TRBC1 (GI:135523) on the forward strand of chromosome 7. The spectra for this peptide are annotated by a fragment of porcine trypsin with similar sequence (LGEHNIDVLEG NEQFINAAK).
Many of the peptides not present in IPI are present in other isoforms or proteins found in the NCBI nonredundant database. We observe a total of 90 such peptides (1938 spectra). See Supplemental Table 1 for the complete list. These cases illustrate the danger of selecting a limited set of “representative” splice forms for a protein database. After removing such annotations, we retain 58,000 novel spectra (6100 peptides). We note that incorrect matches are more likely to be novel peptides, since 80% of the exon graph database is novel sequence. Let us conservatively assume the incorrect matches all fall within the novel peptides. Given a 2.5% false discovery rate across all 1.2 M annotations, we estimate that 28,000 spectra are correctly annotated by novel peptides. These correspond to an estimated 3300 peptides, based on the mean number of spectra per novel peptide. A report of all novel peptides is provided in Supplemental Table 2.
In the remainder of our analysis, we restrict our attention to those novel peptides strongly supported by additional lines of evidence. We find evidence for novel exons (or extensions of known exons) in 16 genes. These instances are supported by sequence homology and by the discovery of one or more peptides in close proximity along the genome. The discovery of translated peptides demonstrates that these sites are indeed exons and not conserved noncoding sequences. See Figure 6 for an example of the evidence for one exon.
Figure 6.
Novel exons are supported by peptide identifications and by sequence homology. Above is a multiple alignment for hypothetical protein sequences from chimp (gi:55639283), rat (gi:62531299), and human (genome translation, similar to PGAM5 gi:20070384). Introns are indicated by colons. The peptides identified from mass spectra are indicated below the protein sequence. The novel 3′ exon is supported by three peptide identifications, as well as >95% amino acid sequence conservation across species.
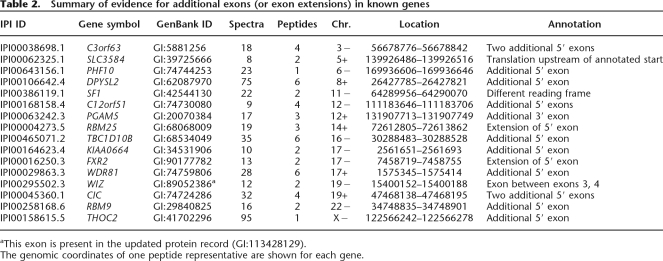
Table 2 summarizes these exon discoveries. While the main purpose of our project is the preliminary annotation of nonannotated or sparsely annotated genomes, the discovery of new exons on the human genome demonstrates the power of the technique. In most cases, the novel translation is immediately upstream of known exons. We note that many of the reference protein sequences are derived from cDNA sequences. The 5′ portions of such sequences are often inferred or absent due to truncation of cDNA. In addition, predicted translation start sites are often incorrect. With the exon graph, we can use mass spectra not only to confirm translation of these genes but to correct their sequence annotations. Supplemental Table 3 reports the peptide hits to these novel exons, as well as peptides from the known exons of the protein. Supplemental Figure 2 illustrates one such case.
Table 2.
Summary of evidence for additional exons (or exon extensions) in known genes
aThis exon is present in the updated protein record (GI:113428129).
The genomic coordinates of one peptide representative are shown for each gene.
Two peptides were observed that fall within splicing factor 1 (GI:42544130) but not in the annotated reading frame. These peptides are of particular interest since they fall within one of the genomic regions selected by the ENCODE project (ENCODE Project Consortium 2004).
Alternative splicing
Evidence for alternative splicing normally comes from mRNA sequencing projects, which may include prespliced or contaminating sequences. Mass spectrometry data can confirm the presence of specific isoforms in a sample at the protein level. Of our peptide matches, ∼25% span at least one putative intron. Overlapping exon predictions and EST alignments can produce unreasonably short exons in the database; therefore, we discard peptides undergoing two splice events within 15 bp of each other.
We examined our search results for evidence of alternative splicing. We consider all splice donors and splice acceptors that have multiple partners. We ignore matches where the splice boundaries are not part of a known protein, or where the peptide covers six or fewer base pairs on either side of the intron. We highlight a total of 40 instances of alternative splicing in this way. We report these events in Supplemental Table 4.
In 24 of these instances, only one of the two isoforms is present in the IPI database. As s conservative filter, we report such splice junctions only if they are supported by EST evidence and/or supported by sequences in the NCBI nonredundant database.
Polymorphisms
Each known coding SNP produces a “bulge” in the exon graph, where a peptide sequence may not match the genomic sequence. A total of 308 such polymorphisms in known genes were evidenced by at least two spectrum hits (see Supplemental Table 5). For 94 of these cases, both alleles of the SNP were observed. In addition, 221 sites were observed where the observed peptide matches the genomic sequence, rather than the protein from the IPI database. These sites may correspond to SNPs, or simply to sequencing errors. We note that many protein records are derived from error-prone sources such as single-pass cDNA sequencing.
Hypothetical proteins
Many protein records in the IPI database are derived from high-throughput cDNA experiments or computational gene predictions. Identification of peptides from these proteins serve as confirmation that the locus in question is, in fact, a protein-coding gene. We examined all search results that correspond to proteins with annotations of the form “hypothetical protein” or “putative protein.” We disregarded any search hits that also match “nonhypothetical” proteins, due to either exons shared with other proteins or multiple occurrences of the peptide within the database. The search results confirm many hypothetical proteins. A total of 224 proteins are matched by a minimum of five spectra from at least two distinct peptides. We omit from this list any sequence present in RefSeq (Pruitt et al. 2005) with an annotation other than “REFSEQ PREDICTED” or “REFSEQ MODEL.” Supplemental Table 6 summarizes the results. This may be the first confirmation of these protein sequences at the level of translation. Supplemental Figure 3 shows coverage of one such protein.
Refining gene predictions
Here we address the question: Can de novo gene finding be improved by incorporating evidence from mass spectrometry? Earlier research has demonstrated the effectiveness of incorporating additional lines of evidence, such as comparative genomics, to improve gene prediction (Korf et al. 2001). By searching mass spectra against our database of putative proteins, we accumulate evidence supporting putative exons and introns. When predicting genes, GeneID first identifies putative exons, then assembles the exons into a collection of genes. We rescore the predicted exons before gene assembly in an effort to improve the accuracy of gene prediction. We boost exon and intron scores based on the number of spectra matched by corresponding peptides and based on the quality of these matches (see Methods).
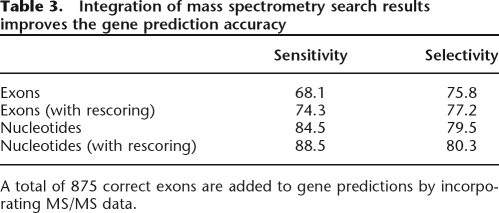
We ran GeneID on the genomic intervals containing 1386 protein-coding genes. We selected genes for which one or more peptides were mapped to the coding region, and for which a single splice isoform was known (from the IPI database). We then rescored all predicted exons by incorporating peptide matches from our database search. The sensitivity and selectivity of gene assembly improved (Table 3), with a gain of 863 correctly identified exons. The improvements are greatest for proteins that are well sampled (data not shown). We also note that since we examine a broad selection of genes, including 100 that span >100,000 bp, accuracy on this corpus may be lower than on other test sets. Figure 7 shows an example of a gene prediction improved by this method.
Table 3.
Integration of mass spectrometry search results improves the gene prediction accuracy
A total of 875 correct exons are added to gene predictions by incorporating MS/MS data.
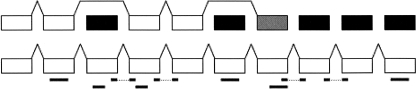
Figure 7.
Diagram of gene prediction results for RFC4 (IPI00017381.1), before (above) and after incorporation of MS/MS results. Correctly predicted exons are shown in white; missed exons, in black. A partially correct exon is shown in gray. Peptide identifications are indicated below their exons (and spanned introns). After exons are rescored using the identified peptides, the full gene is predicted correctly. (Figure not to scale.)
In a few cases (20 genes), predictions worsened after rescoring. The peptide annotations used for these genes appear to be correct. In most cases, an incorrect exon (which overlaps the true exon) was boosted and selected for the final gene prediction. One instance of a peptide mapped to an incorrect splice boundary was also observed. Further work will focus on improved incorporation of MS/MS data, and integration of MS/MS search results alongside other data that can corroborate exons (ESTs and comparative genomics). We anticipate that refinement of the algorithm as well as acquisition of additional spectra will improve results.
Discussion
Delineating the protein-coding genes within a eukaryotic genome remains a complex and labor-intensive process. To cite one example, a human-curated annotation of the human X chromosome required an estimated 15,000 person-hours (Harsha et al. 2005), much of which was spent resolving the set of coding regions. Because automated annotations are the foundation that biologists later build upon, high-throughput methods to generate and refine annotations are needed. This study demonstrates that with a few mass spectrometry experiments, automated analysis can recapture many of the gene annotations that have been made by painstaking efforts. Even on the extensively studied human genome, we discover genes and exons that have not yet been deposited in sequence databases. The majority of our data were drawn from two tissue sources (kidney cells and blood plasma). Consideration of other tissues or enrichment for specific organelles will surely expand our picture of the proteome. On a less thoroughly annotated genome, we expect to see a readout of many more novel genes.
The exon graph is a compact representation of protein splice isoforms and polymorphisms. We observe a near 10-fold reduction in database size between dbEST and the exon graph. We emphasize that this is difficult to accomplish with a typical database, stored in FASTA format. Enumeration of all protein sequences greatly increases search time and creates confusion when matches to dozens of “records” are explained by one gene. Many databases sidestep the problem by including one or two representative sequences for each protein, but this approach carries omits isoforms and polymorphisms. Algorithmic improvements are one way to reduce redundancy from linear protein databases (Edwards and Lippert 2004). We believe that, if available in a standard vendor- and tool-independent file format, exon graph databases may be of general interest to proteomics researchers.
We used two data sources that complement each other to construct the exon graph. An advantage of the EST evidence is that it includes evidence for introns. Short exons, or exons with unusual hexamer count, are difficult to identify de novo but may be covered by ESTs. A limitation of EST evidence is that ESTs may not be available for all genes, and may not cover the 5′ portion of a gene. Many genes are transcribed only in certain tissues or under certain conditions and may never have been captured as ESTs. Another drawback of EST data is the presence of unprocessed and truncated transcripts, as well as genomic contaminants. Exon predictions have the advantage that they explicitly indicate reading frame. Database construction proceeds from putative exons and introns, independent of any specific exon prediction method. We are working to integrate other signals including the output from multiple gene finding programs, evolutionarily conserved regions, etc.
Our results include 40 instances of alternative splicing. We emphasize that we have highlighted only those instances where two splicing events are observed at the same locus. These results directly confirm both splice events. Many other peptide identifications are unique to splice isoforms that are not considered standard, giving indirect evidence of alternative splicing. It is notable that many splice isoforms differ by the inclusion of a single amino acid. These are cases where two splice donor (or acceptor) sites are present, separated by 3 bp. Some isoforms of biological significance differ by presence or absence of a single amino acid (Tadokoro et al. 2005).
Fully characterizing splice events from tryptic peptides gives rise to a phasing problem which may be avoided by top-down mass spectrometry of complete proteins (Roth et al. 2005). Mass spectrometry can reliably demonstrate the presence of protein isoforms, but confirming their absence is problematic (Godovac-Zimmermann et al. 2005). Sequence-based methods remain important, particularly for splice events that take place in the untranslated region of genes.
Our focus in this article is on cataloging coding exons and splice events. We note that mass spectrometry can measure other types of information that are invaluable for annotation of genes. These include post-translational modifications (Jensen 2006), proteolytic cleavages (e.g., of signal peptides) (N. Gupta, S. Tanner, N. Jaitly, J. Adkins, M. Lipton, R. Edwards, M. Romine, A. Osterman, V. Bafna, R. Smith, in prep.), subcellular localization (Dunkley et al. 2006), and relative protein expression levels between tissues (Lill 2003). These topics are a subject of ongoing research. Our search did not consider post-translational modifications explicitly. Some modified peptides were annotated with a sequence with the same mass as the true (modified) peptide. For example, the putative peptide ASVVAVSDGVIK matches the N-terminally acetylated peptide from CFL1 (A+42SGVAV SDGVIK). The putative peptide DELHIVEAEAVYYKGSPIK matches a modified peptide DELHIVEAEAM+16NYKGSPIK from IPI00455423.1 (similar to NPM1). Using other algorithms developed in our laboratory (Tsur et al. 2005), we are searching these same data sets for known and unknown post-translational modifications. Similar studies are underway for bacterial genomes (N. Gupta, S. Tanner, N. Jaitly, J. Adkins, M. Lipton, R. Edwards, M. Romine, A. Osterman, V. Bafna, R. Smith, in prep.).
We argue that high-throughput proteomics experiments should accompany each genome sequencing project. Mass spectrometry is a practical technique for annotating protein-coding regions. The search is able to tolerate a substantial overhead of “noise” in exon predictions. In addition, the technique is orthogonal to standard transcript-level methods such as cDNA sequencing. Mass spectrometry complements other experimental methods. With recent advances in instrumentation, the data volume we consider in this article can be produced in 10 instrument-weeks with two person-weeks of labor. Scaling up mass spectrometry experiments to help annotate a large portion of proteomes is an attractive prospect at feasible cost.
Acknowledgments
S.T. is supported by NSF IGERT training grant DGE0504645. This research was supported in part by NIH (RR016522-04A1), and by the UCSD FWGrid Project, NSF Research Infrastructure Grant Number EIA-0303622. Part of this investigation was supported using the computing facility made possible by the Research Facilities Improvement Program Grant Number C06 RR017588 awarded to the Whitaker Biomedical Engineering Institute, and the Biomedical Technology Resource Centers Program Grant Number P41 RR08605 awarded to the National Biomedical Computation Resource, UCSD, from the National Center for Research Resources, National Institutes of Health.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5646507
References
- Aebersold R., Mann M., Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Aho A., Corasick M., Corasick M. Efficient string matching: An aid to bibliographic search. Commun. ACM. 1975;18:333–340. [Google Scholar]
- Bafna V., Edwards N., Edwards N. SCOPE: A probabilistic model for scoring tandem mass spectra against a peptide database. Bioinformatics. 2001;17:13–21. doi: 10.1093/bioinformatics/17.suppl_1.s13. [DOI] [PubMed] [Google Scholar]
- Blanco E., Parra G., Guigó R., Parra G., Guigó R., Guigó R. Current Protocols in Bioinformatics. John Wiley & Sons Inc.; New York: 2002. Using GeneID to identify genes. Unit 4.3. [Google Scholar]
- Boguski M.S., Tolstoshev C.M., Bassett D.E., Jr., Tolstoshev C.M., Bassett D.E., Jr., Bassett D.E., Jr. Gene discovery in dbEST. Science. 1993;265:1993–1994. doi: 10.1126/science.8091218. [DOI] [PubMed] [Google Scholar]
- Carlton J.M., Angiuoli S.V., Suh B.B., Kooij T.W., Pertea M., Silva J.C., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., Angiuoli S.V., Suh B.B., Kooij T.W., Pertea M., Silva J.C., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., Suh B.B., Kooij T.W., Pertea M., Silva J.C., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., Kooij T.W., Pertea M., Silva J.C., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., Pertea M., Silva J.C., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., Silva J.C., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., Allen J.E., Selengut J.D., Koo H.L., Selengut J.D., Koo H.L., Koo H.L., et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- Choudhary J., Blackstock W., Creasy D., Cottrell J., Blackstock W., Creasy D., Cottrell J., Creasy D., Cottrell J., Cottrell J. Interrogating the human genome using uninterpreted mass spectrometry data. Proteomics. 2001;1:651–667. doi: 10.1002/1615-9861(200104)1:5<651::AID-PROT651>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Craig R., Beavis R., Beavis R. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- Creasy D., Cottrell J., Cottrell J. Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics. 2002;2:1426–1434. doi: 10.1002/1615-9861(200210)2:10<1426::AID-PROT1426>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Desiere F., Deutsch E., Nesvizhskii A., Mallick P., King N., Eng J., Aderem A., Boyle R., Brunner E., Donohoe S., Deutsch E., Nesvizhskii A., Mallick P., King N., Eng J., Aderem A., Boyle R., Brunner E., Donohoe S., Nesvizhskii A., Mallick P., King N., Eng J., Aderem A., Boyle R., Brunner E., Donohoe S., Mallick P., King N., Eng J., Aderem A., Boyle R., Brunner E., Donohoe S., King N., Eng J., Aderem A., Boyle R., Brunner E., Donohoe S., Eng J., Aderem A., Boyle R., Brunner E., Donohoe S., Aderem A., Boyle R., Brunner E., Donohoe S., Boyle R., Brunner E., Donohoe S., Brunner E., Donohoe S., Donohoe S., et al. Integration of peptide sequences obtained by high-throughput mass spectrometry with the human genome. Genome Biol. 2004;1:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley T.P.J., Hester S., Shadforth I.P., Runions J., Weimar T., Hanton S.L., Griffin J.L., Bessant C., Brandizzi F., Hawes C., Hester S., Shadforth I.P., Runions J., Weimar T., Hanton S.L., Griffin J.L., Bessant C., Brandizzi F., Hawes C., Shadforth I.P., Runions J., Weimar T., Hanton S.L., Griffin J.L., Bessant C., Brandizzi F., Hawes C., Runions J., Weimar T., Hanton S.L., Griffin J.L., Bessant C., Brandizzi F., Hawes C., Weimar T., Hanton S.L., Griffin J.L., Bessant C., Brandizzi F., Hawes C., Hanton S.L., Griffin J.L., Bessant C., Brandizzi F., Hawes C., Griffin J.L., Bessant C., Brandizzi F., Hawes C., Bessant C., Brandizzi F., Hawes C., Brandizzi F., Hawes C., Hawes C., et al. Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards N., Lippert R., Lippert R. The 4th Workshop on Algorithms in Bioinformatics (WABI) Bergen; Norway: 2004. Sequence database compression for peptide identification from tandem mass spectra. [Google Scholar]
- ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Fermin D., Allen B., Blackwell T., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Allen B., Blackwell T., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Blackwell T., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Adamski M., Xu Y., Ulintz P., Omenn G., States D., Xu Y., Ulintz P., Omenn G., States D., Ulintz P., Omenn G., States D., Omenn G., States D., States D. Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol. 2006;7:R35. doi: 10.1186/gb-2006-7-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea L., Francesco V., Miller J., Turner R., Yao A., Harris M., Walenz B., Mobarry C., Merkulov G., Charlab R., Francesco V., Miller J., Turner R., Yao A., Harris M., Walenz B., Mobarry C., Merkulov G., Charlab R., Miller J., Turner R., Yao A., Harris M., Walenz B., Mobarry C., Merkulov G., Charlab R., Turner R., Yao A., Harris M., Walenz B., Mobarry C., Merkulov G., Charlab R., Yao A., Harris M., Walenz B., Mobarry C., Merkulov G., Charlab R., Harris M., Walenz B., Mobarry C., Merkulov G., Charlab R., Walenz B., Mobarry C., Merkulov G., Charlab R., Mobarry C., Merkulov G., Charlab R., Merkulov G., Charlab R., Charlab R., et al. Gene and alternative splicing annotation with AIR. Genome Res. 2005;15:54–66. doi: 10.1101/gr.2889405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A., Tanner S., Bafna V., Pevzner P., Tanner S., Bafna V., Pevzner P., Bafna V., Pevzner P., Pevzner P. Peptide sequence tags for fast database search in mass spectrometry. J. Proteome Res. 2005;4:1287–1295. doi: 10.1021/pr050011x. [DOI] [PubMed] [Google Scholar]
- Godovac-Zimmermann J., Kleiner O., Brown L.R., Drukier A.K., Kleiner O., Brown L.R., Drukier A.K., Brown L.R., Drukier A.K., Drukier A.K. Perspectives in spicing up proteomics with splicing. Proteomics. 2005;5:699–709. doi: 10.1002/pmic.200401051. [DOI] [PubMed] [Google Scholar]
- Gupta S., Zink D., Korn B., Vingron M., Haas S., Zink D., Korn B., Vingron M., Haas S., Korn B., Vingron M., Haas S., Vingron M., Haas S., Haas S. Strengths and weaknesses of EST-based prediction of tissue-specific alternative splicing. BMC Genomics. 2004;5:72. doi: 10.1186/1471-2164-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsha H., Suresh S., Amanchy R., Deshpande N., Shanker K., Yatish A., Muthusamy B., Vrushabendra B., Rashmi B., Chandrika K., Suresh S., Amanchy R., Deshpande N., Shanker K., Yatish A., Muthusamy B., Vrushabendra B., Rashmi B., Chandrika K., Amanchy R., Deshpande N., Shanker K., Yatish A., Muthusamy B., Vrushabendra B., Rashmi B., Chandrika K., Deshpande N., Shanker K., Yatish A., Muthusamy B., Vrushabendra B., Rashmi B., Chandrika K., Shanker K., Yatish A., Muthusamy B., Vrushabendra B., Rashmi B., Chandrika K., Yatish A., Muthusamy B., Vrushabendra B., Rashmi B., Chandrika K., Muthusamy B., Vrushabendra B., Rashmi B., Chandrika K., Vrushabendra B., Rashmi B., Chandrika K., Rashmi B., Chandrika K., Chandrika K., et al. A manually curated functional annotation of the human X chromosome. Nat. Genet. 2005;37:331–332. doi: 10.1038/ng0405-331. [DOI] [PubMed] [Google Scholar]
- Heber S., Alekseyev M., Sze S., Tang H., Pevzner P.A.,, Alekseyev M., Sze S., Tang H., Pevzner P.A.,, Sze S., Tang H., Pevzner P.A.,, Tang H., Pevzner P.A.,, Pevzner P.A., Splicing graphs and EST assembly problem. Bioinformatics. 2002;18:S181–S188. doi: 10.1093/bioinformatics/18.suppl_1.s181. [DOI] [PubMed] [Google Scholar]
- Jensen O.N. Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A., Kolker E., Aebersold R., Nesvizhskii A., Kolker E., Aebersold R., Kolker E., Aebersold R., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kersey P.J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R., Williams A., Karavidopoulou Y., Birney E., Apweiler R., Karavidopoulou Y., Birney E., Apweiler R., Birney E., Apweiler R., Apweiler R. The international protein index: An integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Korf I., Flicek P., Duan D., Brent M., Flicek P., Duan D., Brent M., Duan D., Brent M., Brent M. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17:S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- Kuster B., Mortensen P., Andersen J.S., Mann M., Mortensen P., Andersen J.S., Mann M., Andersen J.S., Mann M., Mann M. Mass spectrometry allows direct identification of proteins in large genomes. Proteomics. 2001;1:641–650. doi: 10.1002/1615-9861(200104)1:5<641::AID-PROT641>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Leipzig J., Pevzner P., Heber S., Pevzner P., Heber S., Heber S. The Alternative Splicing Gallery (ASG): Bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill J. Proteomic tools for quantitation by mass spectrometry. Mass Spectrom. Rev. 2003;22:182–194. doi: 10.1002/mas.10048. [DOI] [PubMed] [Google Scholar]
- Lu B., Chen T., Chen T. A suffix tree approach to the interpretation of tandem mass spectra: applications to peptides of non-specific digestion and post-translational modifications. Bioinformatics. 2003;19:113–121. doi: 10.1093/bioinformatics/btg1068. [DOI] [PubMed] [Google Scholar]
- Mironov A., Fickett J., Gelfand M., Fickett J., Gelfand M., Gelfand M. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B., Lee C., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. [Google Scholar]
- Omenn G., States D., Adamski M., Blackwell T., Menon R., Hermjakob H., Apweiler R., Haab B., Simpson R., Eddes J., States D., Adamski M., Blackwell T., Menon R., Hermjakob H., Apweiler R., Haab B., Simpson R., Eddes J., Adamski M., Blackwell T., Menon R., Hermjakob H., Apweiler R., Haab B., Simpson R., Eddes J., Blackwell T., Menon R., Hermjakob H., Apweiler R., Haab B., Simpson R., Eddes J., Menon R., Hermjakob H., Apweiler R., Haab B., Simpson R., Eddes J., Hermjakob H., Apweiler R., Haab B., Simpson R., Eddes J., Apweiler R., Haab B., Simpson R., Eddes J., Haab B., Simpson R., Eddes J., Simpson R., Eddes J., Eddes J., et al. Overview of the hupo plasma proteome project: Results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- Parra G., Blanco E., Guigó R., Blanco E., Guigó R., Guigó R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D., Pappin D., Creasy D., Cottrell J., Pappin D., Creasy D., Cottrell J., Creasy D., Cottrell J., Cottrell J. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R., Tatusova T., Maglott D.R., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resing K., Meyer-Arendt K., Mendoza A., Aveline-Wolf L., Jonscher K., Pierce K., Old W., Cheung H., Russell S., Wattawa J., Meyer-Arendt K., Mendoza A., Aveline-Wolf L., Jonscher K., Pierce K., Old W., Cheung H., Russell S., Wattawa J., Mendoza A., Aveline-Wolf L., Jonscher K., Pierce K., Old W., Cheung H., Russell S., Wattawa J., Aveline-Wolf L., Jonscher K., Pierce K., Old W., Cheung H., Russell S., Wattawa J., Jonscher K., Pierce K., Old W., Cheung H., Russell S., Wattawa J., Pierce K., Old W., Cheung H., Russell S., Wattawa J., Old W., Cheung H., Russell S., Wattawa J., Cheung H., Russell S., Wattawa J., Russell S., Wattawa J., Wattawa J., et al. Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal. Chem. 2004;76:3556–3568. doi: 10.1021/ac035229m. [DOI] [PubMed] [Google Scholar]
- Roth M.J., Forbes A.J., Boyne M.T.N., Kim Y.-B., Robinson D.E., Kelleher N.L., Forbes A.J., Boyne M.T.N., Kim Y.-B., Robinson D.E., Kelleher N.L., Boyne M.T.N., Kim Y.-B., Robinson D.E., Kelleher N.L., Kim Y.-B., Robinson D.E., Kelleher N.L., Robinson D.E., Kelleher N.L., Kelleher N.L. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Mol. Cell. Proteomics. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadygov R., Yates J., Yates J. A hypergeometric probability model for protein identification and validation using tandem mass spectral data and protein sequence databases. Anal. Chem. 2003;75:3792–3798. doi: 10.1021/ac034157w. [DOI] [PubMed] [Google Scholar]
- Tabb D., Smith L., Breci L., Wysocki V., Lin D., Yates J., Smith L., Breci L., Wysocki V., Lin D., Yates J., Breci L., Wysocki V., Lin D., Yates J., Wysocki V., Lin D., Yates J., Lin D., Yates J., Yates J. Statistical characterization of ion trap tandem mass spectra from doubly charged tryptic peptides. Anal. Chem. 2003;75:1155–1163. doi: 10.1021/ac026122m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro K., Yamazaki-Inoue M., Tachibana M., Fujishiro M., Nagao K., Toyoda M., Ozaki M., Ono M., Miki N., Miyashita T., Yamazaki-Inoue M., Tachibana M., Fujishiro M., Nagao K., Toyoda M., Ozaki M., Ono M., Miki N., Miyashita T., Tachibana M., Fujishiro M., Nagao K., Toyoda M., Ozaki M., Ono M., Miki N., Miyashita T., Fujishiro M., Nagao K., Toyoda M., Ozaki M., Ono M., Miki N., Miyashita T., Nagao K., Toyoda M., Ozaki M., Ono M., Miki N., Miyashita T., Toyoda M., Ozaki M., Ono M., Miki N., Miyashita T., Ozaki M., Ono M., Miki N., Miyashita T., Ono M., Miki N., Miyashita T., Miki N., Miyashita T., Miyashita T., et al. Frequent occurrence of protein isoforms with or without a single amino acid residue by subtle alternative splicing: The case of gln in drpla affects subcellular localization of the products. J. Hum. Genet. 2005;50:382–394. doi: 10.1007/s10038-005-0261-9. [DOI] [PubMed] [Google Scholar]
- Tanner S., Shu H., Frank A., Wang L., Zandi E., Mumby M., Pevzner P., Bafna V., Shu H., Frank A., Wang L., Zandi E., Mumby M., Pevzner P., Bafna V., Frank A., Wang L., Zandi E., Mumby M., Pevzner P., Bafna V., Wang L., Zandi E., Mumby M., Pevzner P., Bafna V., Zandi E., Mumby M., Pevzner P., Bafna V., Mumby M., Pevzner P., Bafna V., Pevzner P., Bafna V., Bafna V. Inspect: Fast and accurate identification of post-translationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- Tsur D., Tanner S., Zandi E., Bafna V., Pevzner P., Tanner S., Zandi E., Bafna V., Pevzner P., Zandi E., Bafna V., Pevzner P., Bafna V., Pevzner P., Pevzner P. Identification of post-translational modifications via blind search of mass-spectra. Nat. Biotechnol. 2005;23:1562–1567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yates J., Eng J., McCormack A., Eng J., McCormack A., McCormack A. Mining genomes: Correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 1995a;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- Yates J., Eng J., McCormack A., Schieltz D., Eng J., McCormack A., Schieltz D., McCormack A., Schieltz D., Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995b;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]