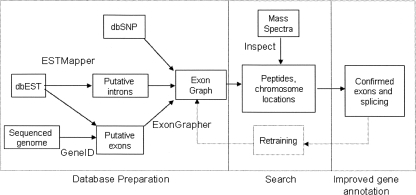
Figure 1.
Overview of the workflow for genome annotation through mass spectrometry. The exon graph database is constructed without reference to prior annotations of the genome. Putative exons and exon-pairs are generated through EST alignment and de novo predictions; homology maps are another potential source. Peptide matches identify the true exons (and introns) among the gene predictions