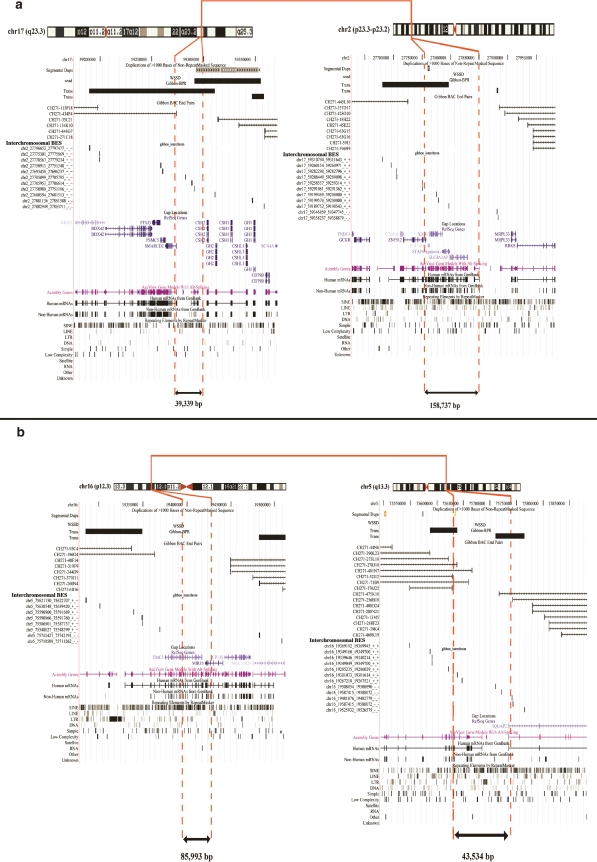
Figure 3.
Molecular refinement of breakpoints by paired-end-sequence mapping. Regions that were confirmed both experimentally and computationally were further refined by consideration of discordant and concordant Nomascus leucogenys BES mapped to the human genome. Two translocation intervals are depicted by incorporating data into the UCSC Browser. (a) 17q23.3 to 2p23.3 rearrangement breakpoints and (b) 16p12.3 to 5q13.3 rearrangement. Interchromosomal NLE BES pairs (interchromosomal BES with coordinates to another chromosome) delineate the location of the breakpoint region (black bars), while concordant NLE BES (Gibbon BAC end pairs) further define the interval of the rearrangement breakpoints (red dotted lines). All underlying BES mapping data are available at http://humanparalogy.gs.washington.edu.