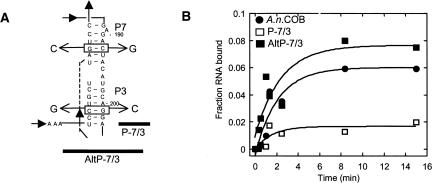
FIGURE 4.
Mutations in P7 and P3 negatively affect binding. (A) Location of the mutations. Only the P7/3 helix is shown. P-7/3 has changes in P7 (C192G) and P3 (G201C) that disrupt base-pairing in the middle of both stems. Compensatory mutations in AltP-7/3 restore base-pairing in both helices. (B) Association kinetics for A.n. COB pre-RNA, P-7/3 and AltP-7/3. Internally labeled RNA was incubated with 4 nM protein, and at various times a molar excess of unlabeled A.n. COB pre-RNA was added to stop binding and the reactions filtered on nitrocellulose. The data were fit to a single exponential to obtain k obs. For three independent experiments, the k obs were: A.n. COB pre-RNA: 0.66 (±0.17) min−1, P-7/3: 0.31 (±0.1) min−1, and AltP-7/3: 0.56 (±0.17) min−1. The k obs at 4 nM I-AniI is essentially the same as measured previously [0.63 (±0.03) min−1; Solem et al. 2002].