Figure 4. Transduction Mediated by HERV-KCON Gag-PR-Pol and Genomes Results in Stable Proviral Integration.
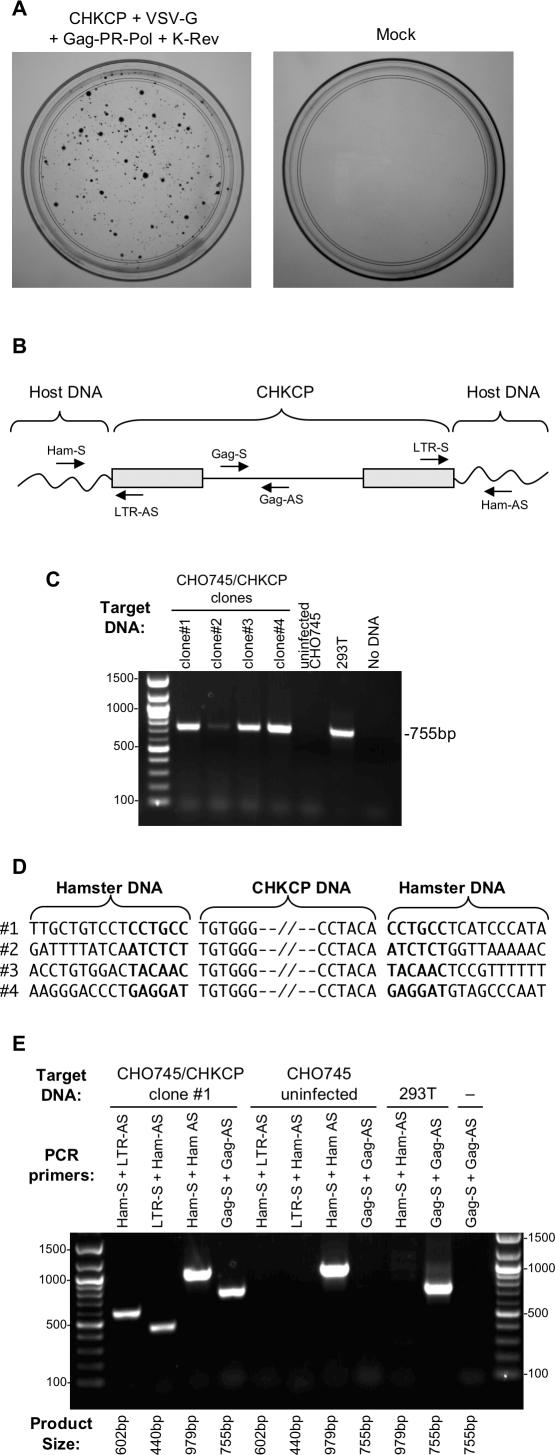
(A) Puromycin-resistant colonies of 293T cells infected with either VSV-G pseudotyped (left) or Env defective (right) virions carrying the CHKCP genome. Infected 293T cells were selected in 0.5 μg/ml puromycin for 2 wk and then fixed and stained to reveal colonies of viable cells. Data are representative of at least three experiments.
(B) Experimental strategy for detection of HERV-KCON proviruses in CHO745 cells using PCR primers targeted to HERV-KCON Gag and LTR sequences, or flanking hamster DNA sequences.
(C) PCR amplification of HERV-KCON gag DNA using Gag-S and Gag-AS primers in four expanded clones of puromycin-resistant CHO745 cells transduced with CHKCP-containing HERV-KCON(VSV-G) particles.
(D) Nucleotide sequences at the 5′ and 3′ ends of integrated CHKCP proviral DNA, revealing six nucleotide duplicated sequences at the CHKCP integration sites.
(E) Verification of the presence and absence of an integrated provirus and the empty preintegration site in CHKCP-transduced and naïve CHO745 cells using combinations of HERV-K and hamster DNA targeted PCR primers (see [B] for primer design strategy). DNA templates and PCR primer pairs used are indicated above each lane, and the expected PCR product size is given below each lane. A representative analysis of a single CHKCG-carrying CHO745 cell clone is shown; similar results were obtained with two additional clones. Uninfected CHO745 cells and human 293T cells serve as controls.