Figure 1.
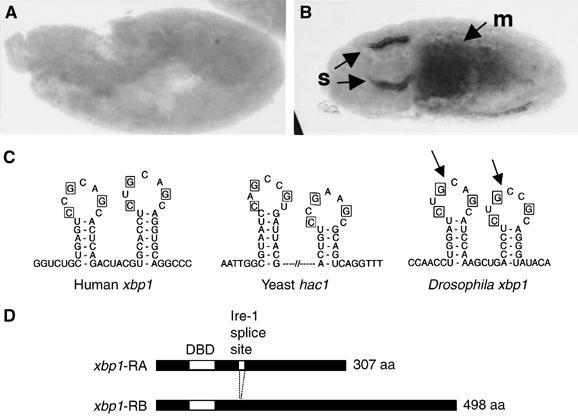
Structure and expression of Drosophila xbp1. (A, B) In situ hybridization against xbp1 mRNA in embryos. (A) A 6 h-old embryo shows little xbp1 mRNA. (B) A 12 h embryo with arrows pointing to high xbp1 mRNA in salivary glands (s) and the midgut (m). (C) A sequence comparison between the xbp1 genes of humans, Drosophila and the yeast hac1 shows a conserved stem–loop structure that is a target of Ire-1-mediated unconventional splicing. The bases required for Ire-1 recognition (in boxes) are conserved in the Drosophila xbp1 gene. The Ire-1 cleavage sites are marked with arrows. (D) The two predicted xbp1 isoforms. After Ire-1-mediated splicing, a frameshift in xbp1 translation occurs, converting a 307 aa protein into a 498 aa protein. The first white box indicates the DNA-binding domain (DBD). The second white box (smaller one) indicates the putative Ire-1 splice site.
