Figure 2.
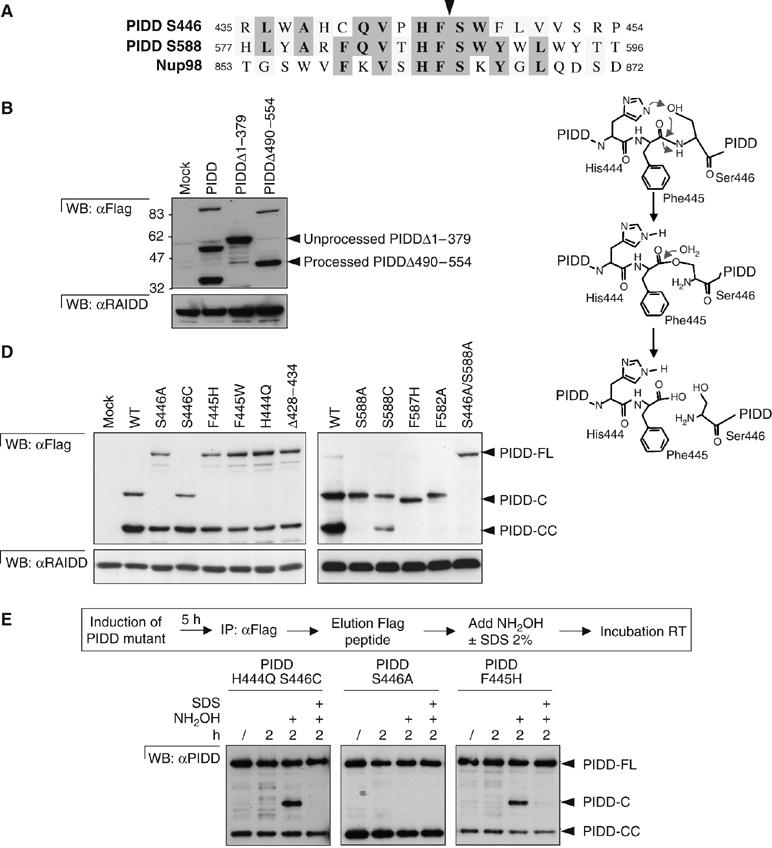
PIDD undergoes auto-processing at Ser446 and Ser588. (A) Alignment of amino-acid sequences surrounding the two PIDD cleavage sites together with the Nup98 cleavage site. The arrow indicates the cleavage site. (B) Deletion mutants of PIDD (Δ1–379 and Δ490–554) or the full-length protein were stably expressed in HEK293T cells and processing was analyzed by Western blotting. (C) Proposed mechanism for the autoproteolytic process of PIDD at the PIDD-C cleavage site, based on the mechanism proposed by Rosenblum and Blobel (1999). An identical mechanism is also proposed for the PIDD-CC cleavage site. (D) PIDD-FL containing mutations in amino acids preceding or following the PIDD-C (left panel) or PIDD-CC (right panel) cleavage sites was analyzed for its capacity to auto-process. (E) In vitro processing of inactive H444Q/S446C, S446A and F445H mutants induced by the nucleophile NH2OH. Expression of the Flag-tagged PIDD mutants was induced by doxocycline treatment of HEK293Trex during 5 h and purified on an anti-Flag affinity column. Immunoprecipitates were eluted using Flag peptides. The various purified PIDD mutants were then incubated with NH2OH in the presence or absence of denaturing SDS. PIDD cleavage was analyzed by Western blotting using the monoclonal anti-PIDD antibody.
