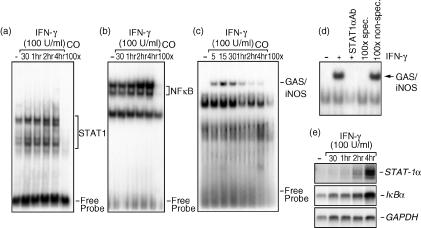
Figure 3.
Nuclear translocation of signal transducer and activator of transcription 1α (STAT1α) and nuclear factor-κB (NF-κB) transcription factors in interferon-γ (IFN-γ)-stimulated J774 macrophages. Nuclear extracts from cells either left untreated or stimulated with IFN-γ (100 U/ml) for different time-periods (0–4 hr) were incubated with a [γ-32P]-labelled STAT1α (consensus sequence) (a), NF-κB (b), or STAT1α [IFN-γ-activated site (GAS)/inducible nitric oxide synthase (iNOS) sequence] (c) probe and were subjected to electrophoretic mobility shift assay (EMSA) analysis. The last lane in each panel represents nuclear extracts from IFN-γ-treated cells incubated with an excess of unlabelled oligonucleotide in order to evaluate band specificity. (d) For supershift assays, nuclear extracts from cells stimulated with IFN-γ (1 hr) were or were not incubated with a specific antibody against STAT1α for 1 hr before EMSA. Binding specificity was tested by adding, to nuclear extracts from 1-hr IFN-γ-treated cells, a 100-fold molar excess of either a cold GAS/iNOS oligonucleotide or a non-specific Oct-2A probe. (e) Macrophages were stimulated with IFN-γ (100 U/ml) for different time-periods (0–4 hr). Following RNA extraction, STAT1α and IκBα gene expression were determined using Northern blot analysis. Equal RNA loading was confirmed by hybridization with a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA probe. These results are representative of one of three separate experiments.