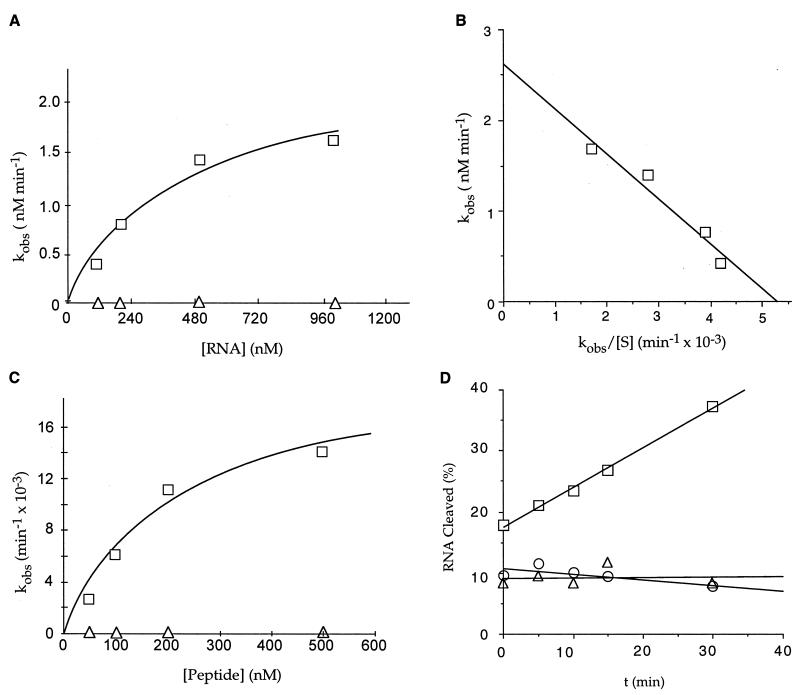
Figure 2.
Kinetics of RNA cleavage by the finger peptide. Multiple- and single-turnover kinetics were performed with the (UGGUGGGCAAUGGGCGUGUU) RNA substrate. (A) Multiple-turnover kinetics for RNA cleavage by the dimeric peptide (□) and zinc-coordinated peptide (▵). The curve represents a nonlinear least-squares fit of the data to the Michaelis–Menten equation. (B) Eadie-Hofstee plot of multiple-turnover kinetics for the dimeric peptide, where [S] is expressed in nM. The slope of the line is equal to −Km, the y intercept is equal to Vmax, and kcat = Vmax/[Et]. The multiple-turnover Km and kcat for the best-fit curve were, respectively, 494 ± 58 nM and 0.026 ± 0.003 min−1. (C) Single-turnover kinetics for the dimeric peptide (□) and the zinc-coordinated peptide (▵). Plotting the kobs as a function of peptide concentration results in a hyperbolic curve with the Km equal to the peptide concentration at half-maximum rate and the kcat equal to the horizontal asymptote. The calculated values for the single-turnover Km and kcat from the best-fit curve were, respectively, 209 ± 64 nM and 0.02 ± .003 min−1. (D) RNA cleavage activity for the reduced (▵) and dimeric (□) forms of the ZFY-6 peptide and the C5,8A peptide mutant (○). The RNA cleavage activity for the reduced form of the peptide was determined by preincubating the peptide and RNA in 20 mM sodium acetate (pH 4) for 30 min and adjusting the pH of the reaction to 7 with 20 mM sodium carbonate (pH 9) for the digestion reaction. The percent RNA cleaved is equal to [product]/[total substrate] × 100.